The Y-dna passes down virtually unchanged from father to son forever, with few mutations.
So it is very specific in tracing our fathers. Whereas Mtdna may be identical to that of his or
her direct maternal ancestor a dozen or so generations ago, but after that, no guarantee.
Because the mutation rate in Mtdna is ten times higher than in nuclear DNA,
this because Mtdna are subject to damage from reactive oxygen
molecules released as a byproduct during OXPHOS. In addition,
Mtdna also lacks the DNA repair mechanisms found in the nucleus.
The Genus and species to which all modern human beings (Homo-sapiens), belong and to which are attributable fossil remains of humans in Africa, from 400,000 years ago or more. (In Qesem Cave near Rosh Haayin, in central Israel, human teeth were found indicating the existence of modern man (Homo sapiens) in Israel as early as about 400,000 years ago; thus proving that modern man is much older than 400,000 years). Homo sapiens are distinguished from other animals and from earlier humanoid species by characteristics and habits such as bipedal stance and gait, brain capacity averaging about 82 cubic inches, high forehead, small teeth and jaw, defined chin, construction and use of tools, and the ability to make use of symbols such as used in language and writing. Some of these features were possessed by the immediate ancestor, Homo erectus; but in the aggregate they are characteristic only of Homo sapiens - Us.
Cells are the basic building blocks of all living things. The human body is composed of trillions of cells. They provide structure for the body, take in nutrients from food, convert those nutrients into energy, and carry out specialized functions. Cells also contain the body’s hereditary material and can make copies of themselves.
As you read the genetic material below,and find it hard to believe,please keep this indisputable fact in mind. |
Genes are segments of deoxyribonucleic acid (DNA) that contain the code for a specific protein that functions in one or more types of cells in the body. Genes are contained in chromosomes, which are in the cell nucleus. The nucleus of every normal human cell contains 23 “Pairs” of chromosomes, for a total of 46 chromosomes. Normally, “Each Pair” consists of one chromosome from the MOTHER and one from the FATHER.
There are 22 pairs of nonsex (autosomal) chromosomes and “One” pair of sex chromosomes. Paired nonsex chromosomes are, for practical purposes, identical in size, shape, and position and number of genes. Because each member of a pair of nonsex chromosomes contains one of each corresponding gene, there is in a sense a “Backup” for the genes on those chromosomes. The 23rd pair is the sex chromosomes (X (mother) and Y (father). In normal people the (nonsex) gene from each parent is the “SAME”. In a “Mutated” gene (the copies of a gene differ from each other), they are known as “ALLELES”. In a “MUTATED” gene: the ancestral allele (Normal allele) of the SNP is called (the A-allele) and is present in the worldwide population of humans. That means sometime during human history, a mutation in one of the gene parts (mother or father) lead to the emergence of the “derived allele” (called the T-allele).
Throughout this page and others, you will see White people who have the SAME genes as the original Humans - Black People. That is because White People are merely the Albinos of Black People, (in the case of Europeans, they are the Albinos of the Dravidians of India), and thus have the very SAME genes, EXCEPT that some of their genes have been damaged in the places that effect Skin Color, Hair Color, and Eye Color. People of ALL phenotypes: (that is the observable characteristics or traits of an organism - which is the physical expression of it's genes), produce Albinos.
(There is three more types of Albinism: (OCA5, OCA6, OCA7)
 |
 |
 |
 |
 |
European Albinos insist that they are not Albinos because they don't have White Hair, Red Eyes, and Bad Eyesight. This is their denial mechanism: they are mostly OCA2 Albinos, so they point to the most severe type of Albinism, which is OCA1: and claim that they are not Albinos because they are not OCA1. Note the Redhead Albinos above and below. The Roman historian Cornelius Tacitus describes the newly arrived in Europe from Central Asia Germanic people: 4. For my own part, I agree with those who think that the tribes of Germany are free from all taint of intermarriages with foreign nations, and that they appear as a distinct, unmixed race, like none but themselves. Hence, too, the same physical peculiarities throughout so vast a population. All have (White Skin) fierce blue eyes, red hair, huge frames, fit only for a sudden exertion. They are less able to bear laborious work. Heat and thirst they cannot in the least endure; to cold and hunger their climate and their soil inure them. |
 |
 |
 |
As is normal in all Mulatto Societies; the Mulattoes equate superiority with the people and culture in charge and in power. Therefore it is no surprise that the Chinese equate the palest skin, the roundest eyes, and the pointiest noses, with the most desirable phenotype. The only problem is that the overwhelming number of Mongol people are Brown or Yellow skinned, with low nose bridges/broad noses and slanty eyes. And so it appears that what the west once thought was the inscrutable nature of the Mongol, was merely the Mongol Race confusion.
Newsstory: South China Morning Post – Sep. 2015,
Like opium dens, sedan chairs and bat-winged junks, women with bound feet were once stereotypical to China. Deliberately crippled to conform to ideals of beauty, these strange, pathetic creatures - to Western eyes - embodied the mysterious ways of the East. Early travel accounts describe the “alluring” manner in which Chinese women with bound feet walked, as they gently swayed and tottered, usually with an amah (maid) on each arm for support. Physiological reasons for this “attractive” faltering gait were never seriously questioned by casual observers.
Carefully sanitized by euphemistic nonsense, foot binding was considered a quaint cultural taste that no outsider could ever fathom. In reality, the underlying appeal was explicitly sexual. Crippled feet required one to walk in a certain mincing manner to avoid toppling over; as a result, it was believed, the inner thigh and pelvic muscles became unusually tight. Thus, more lurid thought processes went, the smaller the bound feet, the stronger the vaginal muscles would be during lovemaking. Adult human feet reduced to 10cm-long stumps – the fabled “golden lily feet” – were the most prized.
At a time when most Chinese people existed only a few rice bowls away from starvation, being able to keep economically unproductive women whose only practical functions – due to crippled feet – were decorative, sexual and reproductive, was a powerful status marker. Chinese women – as ever – colluded in this patriarchal oppression, often for the most well-intended reasons. Aspirational mothers of pretty girls from poor families bound their daughters' feet in the hope of attracting a wealthy match, who could extract their offspring from the desperate poverty that had blighted their own horizons.
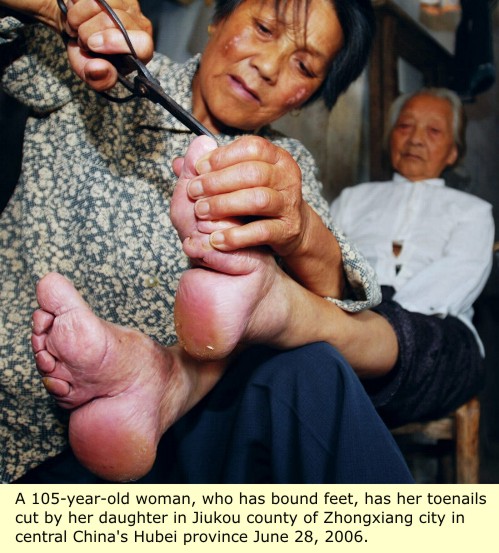 |
In order to keep the deformed bones together, previously bound feet had to be tightly bandaged in a particular manner before the decorative shoes were worn, in much the same way that a boxer’s knuckles are bound with cloth tapes before donning leather gloves. A complicated, time-consuming process, the bandages usually stayed on for days (or even weeks) at a time. When they were eventually undone, the nasty state of the bandages, and the grossly deformed, suppurating feet they covered, can only be imagined – especially in hot weather. The Chinese expression “long and stinking, like granny’s foot-binding cloths …” revoltingly sums it up; the saying is still used to describe overly lengthy, deeply unpleasant personal stories recounted in far too much detail.
Considering the nature of these insular Mongol cultures, it is easy to imagine a Chinese Queen or other "High Status" woman with congenitally deformed feet, and the subsequent need to walk in tiny steps, setting the stage for the "STYLE" of foot binding.
Likewise; note Kublai Khans Albino wife below. Can there be any doubt that she furthered the "STYLE" of wanting to be Albino? |
 |
Like most Mulatto people: I.e. Turk-Arabs, light skinned Hindu's, and other non-Black West Asians, non-Black/White Americans: the Mongol people are terribly Race-confused. As a matter of fact, it appears the Mongol people are starting to follow the European way, and CHOOSING to make MORE Albinos by mating with Albinos. In effect, doubling-down on the DISEASE of Albinism. The BBC Magazine did a very perceptive article titled "A Point Of View: How China sees a multicultural world". << Click here to read the article >>
But regardless of how China sees the Black World, there is the even more important question of how the Black world see China. Modern political and economic relations between China and Africa commenced in the era of Mao Zedong, following the victory of the Chinese Communist Party in the Chinese Civil War. Starting in the 21st century, the modern state of the People's Republic of China has built increasingly strong economic ties with Africa. There are an estimated one million Chinese citizens residing in Africa. Additionally, it has been estimated that 200,000 Africans are working in China. As of 2020, Eswatini is the only African country to have relations with China’s rival Taiwan. Trade between China (PRC) and Africa increased by 700% during the 1990s, and China is currently Africa's largest trading partner. The Forum on China–Africa Cooperation (FOCAC) was established in October 2000 as an official forum to greatly strengthen the relationship. A few Western countries such as the United Kingdom and the United States, have become concerned over the significant political, economic and military roles China is playing in the African continent.
The Dravidian Albinos of East Africa did something that completely changed the course of Human History. We do not know if they were in the first wave of Humans who left East africa or the second. What we do know is that when they crossed Arabia into India, the Black Dravidians stayed in India, while their Albinos headed North into Central Asia, and there they did the unthinkable - they MATED with each other! They broke the human tradition of Albinos never mating with each other, because they knew that they could only produce 100% other Albinos by doing that - with no chance of introducing pigmentation to any of their offspring and gradually returning them to normalcy. Some Dravidians are still at it, see below.
According to evolutionary geneticist Sarah Tishkoff, The (gene) region with the strongest associations was in and around the SLC24A5 gene (an Albinism gene), one variant of which is known to play a role in light skin color in European and some southern Asian populations and is believed to have arisen more than 30,000 years ago. This variant was common in populations in Ethiopia and Tanzania. A note of caution; Sarah is an Albino, and they are known to lie shamelessly about racial issues. Blonde and Red Hair as well as Blue Eyes are a part of the Albinism disease; here they say Blue eyes are only 6,000 - 10,000 years old.
 |
Straight hair - two damaged or recessive allele’s of the "TCHH" gene means straight hair.
Red hair - usually results from a mutation in a gene called MC1R, which codes for the melanocortin-1 receptor.
Blond hair - a single mutation in a long gene sequence called KIT ligand (KITLG) causes people with these genes to have platinum blond, dirty blond or even dark brown hair.
White hair - is caused by a mutated gene called interferon regulatory factor 4, which is important in regulating and producing melanin in the hair.
Blue eyes - a specific mutation within the HERC2 gene, a gene that regulates OCA2 expression, is partly responsible for blue eyes. Other genes implicated in eye color variation are SLC24A4 and TYR.
White skin Mutations - Oculocutaneous Albinism (OCA 1-7) is a group of inherited disorders of melanin biosynthesis characterized by a generalized reduction in pigmentation of hair, skin and eyes. With OCA1A being the most severe type with a complete lack of melanin production throughout life, while the milder forms are OCA1B, OCA2, OCA3, OCA4, OCA5, OCA6, OCA7.
| These Boys (as any sons would) have the exact same genes as their father; except that their Hair Color and Hair Texture genes have been damaged. | This Boy has the exact same genes as his father; except that his Eye Color gene has been damaged. |
ISOGG - International Society of Genetic Genealogy
The root of the Y haplogroup tree is the so-called "Y-Chromosome Adam," the most recent patrineal ancestor of all people living today. He was not the only man living at that time, he simply was the only man with an unbroken male line of descent to the present day. The A haplogroup is thought to have been defined about 60,000 years bp. The BT haplogroup split from the root of the Y haplogroup tree 55,000 years before present (bp), probably in North East Africa. The CF(xDE) haplogroup was the common ancestor of all people who migrated outside of Africa until recent times. The defining mutation occurred 31-55,000 years bp in North East Africa and is still most common in Africa today in Ethiopia and Sudan.
The DE haplogroup appeared approximately 50,000 years bp in North East Africa and subsequently split into haplogroup E that spread to Europe and Africa and haplogroup D that rapidly spread along the coastline of India and Asia to North Asia. The IJ haplogroup characterizes part of the second wave of emigration from Africa that occurred via the Middle East 45,000 years bp and defines two branches I and J that emigrated northwards and eastwards into Europe. The J branch subsequently split again and contributed to the current North African population. The NO haplogroup appeared approximately 35,000-40,000 years bp in a region east of the Aral sea; subsequent branches spread to North Asia (N) and another branch (O) to South Asia via North India.
 |
 |
Haplogroups A & B
Haplogroup A is the non-recombining region of the Y-chromosome (NRY) macrohaplogroup from which all modern paternal haplogroups descend. It is sparsely distributed in Africa, being concentrated among Khoi-san populations in the southwest and Nilotic populations toward the northeast in the Nile Valley. BT is a subclade of haplogroup A; more precisely of the A1b clade.
 |
||
San Bushman of the Kalahari desert |
Berber from the Siwa Oasis of Egypt. 28% of those Berbers are Y-dna Haplogroup A/B. |
 |
 |
|
| A great many of the people at the Siwa Oasis are admixed Turk Mulattoes. | A great many of the people at the Siwa Oasis are admixed Turk Mulattoes. |
 |
ISOGG - International Society of Genetic Genealogy
Y-DNA haplogroup A contains lineages deriving from the earliest branching in the human Y chromosome tree. The oldest branching event, separating A1-P305 and A1-V161, is thought to have occurred about 140,000 years ago. Haplogroups A1-P305, A1a-M31 and A1b1a-M14 are restricted to Africa and A1b1b-M32 is nearly restricted to Africa. The haplogroup that would be named A1b2 is composed of haplogroups B through T. The internal branching of haplogroup A1-V161 into A1a-M31, A1b1, and BT (A1b2) may have occurred about 110,000 years ago. A1-P305 is found at low frequency in Central and West Africa. A1a-M31 is observed in northwestern Africans; A1b1a-M14 is seen among click language-speaking Khoisan populations. A1b1b-M32 has a wide distribution including Khoisan speaking and East African populations, and scattered members on the Arabian Peninsula.
By the definition of haplogroup A as "non-BT", it is almost completely restricted to Africa, though a very small handful of bearers have been reported in Europe and Western Asia. The clade achieves its highest modern frequencies in the Bushmen hunter-gatherer populations of Southern Africa, followed closely by many Nilotic groups in Eastern Africa. However, haplogroup A's oldest sub-clades are exclusively found in Central-Northwest Africa, where it (and by extension the patrilinear ancestor of modern humans) is believed to have originated. Estimates of its time depth have varied greatly, at either close to 190 kya or close to 140 kya in separate 2013 studies, and with the inclusion of the previously unknown "A00" haplogroup to about 270 kya in 2015 studies.
The clade has also been observed at notable frequencies in certain populations in Ethiopia, as well as some Pygmy groups in Central Africa, and less commonly Niger–Congo speakers, who largely belong to the E1b1a clade. Haplogroup E in general is believed to have originated in Northeast Africa, and was later introduced to West Africa from where it spread around 5,000 years ago to Central, Southern and Southeastern Africa with the Bantu expansion. According to Wood et al. (2005) and Rosa et al. (2007), such relatively recent population movements from West Africa changed the pre-existing population Y chromosomal diversity in Central, Southern and Southeastern Africa, replacing the previous haplogroups in these areas with the now dominant E1b1a lineages. Traces of ancestral inhabitants, however, can be observed today in these regions via the presence of the Y DNA haplogroups A-M91 and B-M60 that are common in certain relict populations, such as the Mbuti Pygmies and the Khoisan.
In a composite sample of 3551 African men, Haplogroup A had a frequency of 5.4%. The highest frequencies of haplogroup A have been reported among the Khoisan (combination of khoikhoi who were Nomadic herders of Cattle, Goats, Sheep - pastoralists: and the San who were Nomadic Hunter-Gathers) of Southern Africa, Beta Israel (Real Jews of Ethiopia), and Nilo-Saharans (people who live along the Nile) from Sudan.
Beta Israel, formerly called Falasha also spelled Felasha, now known to be pejorative, Jews of Ethiopian origin. Their beginnings are obscure and possibly polygenetic. The Beta Israel (meaning House of Israel) themselves claim descent from Menilek I, traditionally the son of the Queen of Sheba (Makeda) and King Solomon. At least some of their ancestors, however, were probably local Agau (Agaw, Agew) peoples in Ethiopia who converted to Judaism in the centuries before and after the start of the Christian Era. Although the early Beta Israel remained largely decentralized and their religious practices varied by locality, they remained faithful to Judaism after the conversion of the powerful Ethiopian kingdom of Aksum to Christianity in the 4th century ce, and thereafter they were persecuted and forced to retreat to the area around Lake Tana, in northern Ethiopia.
 |
 |
The Dinka people are a Nilotic ethnic group native to South Sudan, they live mostly along the Nile River - 62% of them are haplogroup A.
 |
Ethopians are 17.8 haplogroup A, Ethopian Jews (Beta Israel) are 40% haplogroup A.
Different Khoisan people are around 45% haplogroup A. (The Khoekhoe or (Khoikhoi) are the traditionally nomadic Pastoralist indigenous population of southwestern Africa. They are grouped with the Hunter-Gatherer San (literally "Foragers") peoples. Thus Khoikhoi-San = Khoisan.
 |
The Nama People (or Nama-Khoe people) are the largest group of the Khoikhoi people, most of whom have largely disappeared as a group, except for the Namas. Many of the Nama clans live in Central Namibia and the other smaller groups live in Namaqualand, which today straddles the Namibian border with South Africa - the Nama people are 64% haplogroup A.
 |
The Nuba peoples are various indigenous ethnic groups who inhabit the Nuba Mountains of South Kordofan state in Sudan, encompassing multiple distinct peoples that speak different languages which belong to at least two unrelated language families - the Nuba people are 46% haplogroup A.
 |
The Shilluk are the fourth largest ethnic group of Southern Sudan, after the Azande and their neighbours the Dinka and Nuer. Their language is called Dhøg Cøllø, dhøg being the Shilluk word for language and mouth. It belongs to the Luo branch of the Western Nilotic subfamily of Nilo-Saharan - Shillup people are 53% haplogroup A: 27% of the Shilluk are haplogroup B.
 |
 |
Haplogroup B (B-M60) is a human Y-chromosome DNA haplogroup common to paternal lineages in Africa. It is a primary branch of the haplogroup BT. B-M60 is common in parts of Africa, especially the tropical forests of West-Central Africa. It was the ancestral haplogroup of not only modern Pygmies like the Baka and Mbuti, but also Hadzabe from Tanzania, who often have been considered, in large part because of some typological features of their language, to be a remnant of Khoisan people in East Africa.
The Nuer people are a Nilotic ethnic group primarily inhabiting the Nile Valley. They are concentrated in South Sudan, and also constitute a majority in Ethiopia's Gambela
Region. They speak the Nuer language, which belongs to the Nilo-Saharan family - 33% of the Nuer people are haplogroup A. 50% of the Nuer people are haplogroup B.
 |
The Hadza, or Hadzabe, are an indigenous ethnic group in north-central Tanzania, living around Lake Eyasi in the central Rift Valley and in the neighboring Serengeti Plateau - 57% of the Hadza are haplogroup B.
 |
Bambuti are pygmy hunter-gatherers, and are one of the oldest indigenous people of the Congo region of Africa. The Bambuti are composed of bands which are relatively small in size, ranging from 15 to 60 people - a little over half of all Pygmies are haplogroup B.
Batwa Pygmies
 |
Zulu are a nation of Nguni-speaking people in KwaZulu-Natal province, South Africa. They are a branch of the southern Bantu and have close ethnic, linguistic, and cultural ties with the Swazi and Xhosa - 20% of Zulu are haplogroup B.
 |
Haplogroup CT is a human Y chromosome haplogroup, defining one of the major paternal lineages of humanity.
Haplogroup CT (P143)
The defining mutations separating CT (all haplogroups except for A and B) are M168 and M294. The site of origin is likely in Africa. Its age has been estimated at approximately 88,000 years old, and more recently at around 100,000 or 101,000 years old.
Men who carry the CT clade have Y chromosomes with the SNP mutation M168, along with P9.1 and M294. These mutations are present in all modern human male lineages except A and B-M60, which are both found almost exclusively in Africa.
The most recent common male line ancestor (TMRCA) of all CT men today probably predated the recent African origin of modern humans, a migration in which some of his descendants participated. He is therefore thought to have lived in Africa before this proposed migration. In keeping with the concept of "Y-chromosomal Adam" given to the patrilineal ancestor of all living humans, CT-M168 has therefore also been referred to in popularized accounts as being the lineage of "Eurasian Adam" or "Out of Africa Adam".
No male in paragroup CT* has ever been discovered in modern populations. This means that all males carrying this haplogroup are also defined as being in one of the several major branch clades. All known surviving descendant lineages of CT are in one of two major subclades, CF and DE. In turn, DE is divided into a predominantly Asia-distributed haplogroup D-CTS3946 and a predominantly Africa-distributed haplogroup E-M96, while CF is divided into an East Asian, Native American, and Oceanian haplogroup C-M130 and haplogroup F-M89, which dominates most non-African populations.
 |
Haplogroup CT had been found in various prehistoric West Asian human fossils that were analysed for ancient DNA, including specimens associated with the Pre-Pottery Neolithic C (1/1; 100%), Neolithic Ganj Dareh Iran (1/2; 50%), Natufian (2/5; 40%), Pre-Pottery Neolithic B (2/7; ~29%), Alföld Linear Pottery (1/1 at two ALP archaeological sites; 100%), Linearbandkeramik (1/2 at Karsdorf LBK archaeological site; 50%) cultures, and some Upper Paleolithic Europeans (Cioclovina1, Kostenki12, Vestonice13). But whether these, or all of them, belong to paragroup CT* or to its branches, is as yet undetermined.
 |
ISOGG
Y-DNA haplogroup C is thought to have arisen shortly after modern humans left Africa. Descendants spread mostly through southern and eastern Asia, but branches developed also in the Americas, islands south of Malaysia, Australia, central Asia and Europe."
Haplogroup C is a major Y-chromosome haplogroup, defined by UEPs M130/RPS4Y711, P184, P255, and P260, which are all SNP mutations. It is one of two primary branches of Haplogroup CF alongside Haplogroup F. Haplogroup C is found in ancient populations on every continent except Africa and is the predominant Y-DNA haplogroup among males belonging to many peoples indigenous to East Asia, Central Asia, Siberia, North America and Australia as well as ancient and modern populations in Europe, the Levant, and Japan. The haplogroup is also found with moderate to low frequency among many present-day populations of Southeast Asia, South Asia, and Southwest Asia.
Haplogroup C (M130)
Haplogroup C (M130, M216) Found in Asia, Oceania, and North America: Haplogroup C1 (F3393/Z1426): Haplogroup C1a (CTS11043): Haplogroup C1a1 (M8, M105, M131) Found with low frequency in Japan: Haplogroup C1a2 (V20) Found with low frequency in Europe, Armenians, Algeria, and Nepal: Haplogroup C1b (F1370, Z16480): Haplogroup C1b1 (AM00694/K281): Haplogroup C1b1a (B66/Z16458): Haplogroup C1b1a1 (M356) Found with low frequency in South Asia, Southwest Asia, and northern China: Haplogroup C1b1a2 (B65): Haplogroup C1b1a2a (B67) Found among Lebbo' people in Borneo, Indonesia: Haplogroup C1b1a2b (F725) Found among Han Chinese (Guangdong, Hunan, and Shaanxi), Dai people (Yunnan), Murut people (Brunei), Malay people (Singapore), and Aeta people (Philippines): Haplogroup C1b1a3 (Z16582) Found with low frequency in Saudi Arabia and Iraq: Haplogroup C1b1b (B68) Found among Dusun people (Brunei): Haplogroup C1b2 (C-Z16582): Haplogroup C1b3 (B477/Z31885): Haplogroup C1b3a (M38) Found in Indonesia, New Guinea, Melanesia, Micronesia, and Polynesia: Haplogroup C1b3b (M347, P309) Found among the indigenous peoples in Australia: Haplogroup C2 (M217, P44) Found throughout Eurasia and North America, but especially among Mongols, Kazakhs, Tungusic peoples, Paleosiberians, and Na-Dené-speaking peoples.
 |
 |
 |
 |
 |
 |
 |
Quote: it is possible that the old Haplogroup C1a2, F and/or E may be those with the oldest presence in Europe.
Upper Paleolithic
Replacement of Neanderthals by early modern humans
It is thought that modern humans began to inhabit Europe during the Upper Paleolithic about 40,000 years ago. Some evidence shows the spread of the Aurignacian culture. From a purely patrilineal, Y-chromosome perspective, it is possible that the old Haplogroup C1a2, F and/or E may be those with the oldest presence in Europe. They have been found in some very old human remains in Europe. However, other haplogroups are far more common among (Current) European males. (The Central Asian R1 & R2).
| Sample Code | Country | Cal BP 95.4% | Culture | Remains | SNP Panel | Sex | mtDNA haplogroup | Y chrom. haplogroup | Genetic Cluster | SNPs covered |
| UstIshim | Russia | 47,480-42,560 | Unassigned | Femur | Shotgun | M | R | K (xLT) | Unassigned | 2,137,615 |
| Oase1 | Romania | 41,640-37,580 | Unassigned | Mandible | Shotgun | M | N | F | Unassigned | 285,076 |
| Kostenki14* | Russia | 38,680-36,260 | Unassigned | Tibia | 3.7M | M | U2 | C1b | Unassigned | 1,774,156 |
| GoyetQ116-1 | Belgium | 35,160-34,430 | Aurignacian | Humerus | 1240k | M | M | C1a | Unassigned | 846,983 |
| Muierii2 | Romania | 33,760-32,840 | Unassigned | Temporal | 3.7M | F | U6 | Unassigned | 98,618 | |
| Paglicci133 | Italy | 34,580-31,210 | Gravettian | Tooth | 1240k | M | U8c | I | Vestonice | 82,330 |
| Cioclovina1 | Romania | 33,090-31,780 | Unassigned | Cranium | 1240k | M | U | CT | Unassigned | 12,784 |
| Kostenki12 | Russia | 32,990-31,840 | Unassigned | Cranium | 3.7M | M | U2 | CT | Unassigned | 61,228 |
| KremsWA3 | Austria | 31,250-30,690 | Gravettian | Cranium | 1240K | M | U5 | Vestonice | 203,986 | |
| Vestonice13 | Czech | 31,070-30,670 | Gravettian | Femur | 3.7M | M | U8c | CT(notIJK) | Vestonice | 139,568 |
| Vestonice15 | Czech | 31,070-30,670 | Gravettian | Femur | 3.7M | M | U5 | BT | Vestonice | 30,900 |
| Vestonice14 | Czech | 31,070-30,670 | Gravettian | Femur | 390k | M | U | Vestonice | 5,677 | |
| Pavlov1 | Czech | 31,110-29,410 | Gravettian | Femur | 3.7M | M | U5 | C1a2 | Vestonice | 57,005 |
| Vestonice43 | Czech | 30,710-29,310 | Gravettian | Femur | 3.7M | M | U | F | Vestonice | 163,946 |
| Vestonice16 | Czech | 30,710-29,310 | Gravettian | Femur | 3.7M | M | U5 | IJK | Vestonice | 945,292 |
| Ostuni2 | Italy | 29,310-28,640 | Gravettian | Femur | 3.7M | F | U2 | Vestonice | 17,017 | |
| GoyetQ53-1 | Belgium | 28,230-27,720 | Gravettian | Fibula | 1240k | F | U2 | Vestonice | 12,567 | |
| Paglicci108 | Italy | 28,430-27,070 | Gravettian | Phalanx | 1240k | F | U2'3'4'7'8'9 | Vestonice | 4,330 | |
| Ostuni1 | Italy | 27,810-27,430 | Gravettian | Tibia | 3.7M | F | M | Vestonice | 369,313 | |
| GoyetQ376-19 | Belgium | 27,720-27,310 | Gravettian | Humerus | 1240k | F | U2 | Vestonice | 25,400 | |
| GoyetQ56-16 | Belgium | 26,600-26,040 | Gravettian | Fibula | 1240k | F | U2 | Vestonice | 9,988 | |
| Malta1 | Russia | 24,520-24,090 | Unassigned | Humerus | Shotgun | M | U | R | Mal'ta | 1439501 |
| ElMiron | Spain | 18,830-18,610 | Magdalenian | Toe | 3.7M | F | U5b | El Mirón | 797,714 | |
| AfontovaGora3 | Russia | 16,930-16,490 | Unassigned | Tooth | 3.7M | F | R1b | Mal'ta | 286,355 | |
| AfontovaGora2 | Russia | 16,930-16,490 | Unassigned | Humerus | Shotgun | M | Mal'ta | 143,751 | ||
| Rigney1 | France | 15,690-15,240 | Magdalenian | Mandible | 1240k | F | U2'3'4'7'8'9 | El Mirón | 35,600 | |
| HohleFels49 | Germany | 16,000-14,260 | Magdalenian | Femur | 390k | M | U8a | I | El Mirón | 63,151 |
| GoyetQ-2 | Belgium | 15,230-14,780 | Magdalenian | Humerus | 1240k | M | U8a | HIJK | El Mirón | 72,263 |
| Brillenhohle | Germany | 15,120-14,440 | Magdalenian | Cranium | 390k | M | U8a | El Mirón | 13,459 | |
| HohleFels79 | Germany | 15,070-14,270 | Magdalenian | Cranium | 390k | M | U8a | El Mirón | 11,211 | |
| Burkhardtshohle | Germany | 15,080-14,150 | Magdalenian | Cranium | 1240k | M | U8a | I | El Mirón | 38,376 |
| Villabruna | Italy | 14,180-13,780 | Epigravettian | Femur | 3.7M | M | U5b2b | R1b1 | Villabruna | 1,215,433 |
| Bichon | Switzerland | 13,770-13,560 | Azilian | Petrous | Shotgun | M | U5b1h | I2 | Villabruna | 2,116,782 |
| Satsurblia | Georgia | 13,380-13,130 | Epigravettian | Petrous | Shotgun | M | K3 | J2 | Satsurblia | 1,460,368 |
| Rochedane | France | 13,090-12,830 | Epipaleolithic | Mandible | 1240k | M | U5b2b | I | Villabruna | 237,390 |
| Iboussieres39 | France | 12,040-11,410 | Epipaleolithic | Femur | 390k | M | U5b2b | Villabruna | 9,659 | |
| Continenza | Italy | 11,200-10,510 | Mesolithic | Cranium | 3.7M | F | U5b1 | Villabruna | 11,717 | |
| Ranchot88 | France | 10,240-9,930 | Mesolithic | Cranium | 1240k | F | U5b1 | Villabruna | 414,863 | |
| LesCloseaux13 | France | 10,240-9,560 | Mesolithic | Femur | 1240k | F | U5a2 | Villabruna | 8,635 | |
| Kotias | Georgia | 9,890-9,550 | Mesolithic | Tooth | Shotgun | M | H13c | J | Satsurblia | 2,133,968 |
| Falkenstein | Germany | 9,410-8,990 | Mesolithic | Fibula | 390k | M | U5a2c | F | Villabruna | 64,428 |
| Karelia | Russia | 8,800-7,950 | Mesolithic | Tooth | Shotgun | M | C1g | R1a1 | Unassigned | 1,754,410 |
| Bockstein | Germany | 8,370-8,160 | Mesolithic | Tooth | 390k | F | U5b1d1 | Villabruna | 21,977 | |
| Ofnet | Germany | 8,430-8,060 | Mesolithic | Tooth | 390k | F | U5b1d1 | Villabruna | 6,263 | |
| Chaudardes1 | France | 8,360-8,050 | Mesolithic | Tibia | 1240k | M | U5b1b | I | Villabruna | 92,657 |
| Loschbour | Luxembourg | 8,160-7,940 | Mesolithic | Tooth | Shotgun | M | U5b1a | I2a1b | Villabruna | 2,091,584 |
| LaBrana1 | Spain | 7,940-7,690 | Mesolithic | Tooth | Shotgun | M | U5b2c1 | C1a2 | Villabruna | 1,884,745 |
| Hungarian.KO1 | Hungarian | 7,730-7,590 | Neolithic | Petrous | Shotgun | M | R3 | I2a | Villabruna | 1,410,303 |
| Motala12 | Sweden | 7,670-7,580 | Mesolithic | Tooth | Shotgun | M | U2e1 | I2a1b* | Unassigned | 1,874,519 |
| BerryAuBac | France | 7,320-7,170 | Mesolithic | Radius | 1240k | M | U5b1a | I | Villabruna | 54,690 |
| Stuttgart | Germany | 7,260-7,020 | Early Neolithic | Tooth | Shotgun | F | T2c1d1 | Unassigned | 2,078,724 |
Note: All dates are obtained as described in Supplementary Information section 1.When an individual has a direct date from an element from the same skeleton it is marked “Direct”, followed by a hyphen to indicate whether the date is obtained by ultrafiltration (“UF”) or without (“NotUF”). If the date is from the archaeological layers, we mark the date type as “Layer”. All the dates were calibrated using IntCal1328 and the OxCal4.2 program29.*We represent Kostenki14 in most analyses by our newly reported 16.1x capture data, but repeat key analyses on the previously reported 2.8x shotgun data30+Mean coverage is computed on the 3.7M SNP targets. |
 |
The Negrito are several ethnic groups of the Australoid race who inhabit isolated parts of Southeast Asia. Their current populations include 12 Andamanese peoples of the Andaman Islands, six Semang peoples of Malaysia, the Mani of Thailand, and the Aeta/Agta and Ati, and 30 other peoples of the Philippines. Genetically, Negritos are the most distant human population from Africans at most loci studied thus far (except for MC1R, which codes for dark skin). The all live in remote areas throughout the islands in the Philippines. (This means that the Negritos were the first wave of modern humans to leave Africa).
The Aeta and other indigenous tribes of the Philippines are Y-dna haplogroups
(C-RPS4Y, K-M9, O-M119, O-M110, and O-M122) at frequencies of 45%.
 |
 |
 |
 |
 |
The history of the Philippines is believed to have begun with the arrival of the first humans using rafts or boats at least 67,000 years ago as the 2007 discovery of Callao Man suggested. Negrito groups were the first inhabitants to settle in prehistoric Philippines. After that, groups of Austronesian (speaking people) later migrated to the islands. Austronesian-speaking peoples, are a large group of various peoples in Taiwan, Island Southeast Asia, Micronesia, coastal New Guinea, Island Melanesia, Polynesia, and Madagascar, that speak the Austronesian languages.
The Chinese sailed around the Philippine Islands from the 9th century onward and frequently interacted with the local Austronesian people. Chinese and Austronesian interactions initially commenced as bartering items. This is evidenced by a collection of Chinese artifacts found throughout Philippine waters, dating back to the 10th century. Minnan peoples started migrating to the Philippines in large numbers from the early 1800s and continue to the present, eventually outnumbering the Cantonese who had always formed the majority Chinese dialect group in the country.
Japanese contact with the Philippine islands began when Japanese traders/merchants first settled in the archipelago during the 12th century AD. In the course of time, shipwrecked Japanese sailors, pirate traders, and immigrants settled in the Philippines and intermarried with the early Filipinos.
Portuguese-born Spanish explorer Ferdinand Magellan conquered Malacca City in 1511 and reached Maluku Islands in 1512. In 1578, the Castilian War erupted between the Christian Spaniards and Muslim Bruneians over control of the Philippine archipelago. The Philippines was never profitable as a colony during Spanish rule, and the long war against the Dutch from the West, in the 17th century together with the intermittent conflict with the Muslims in the South and combating Japanese Wokou piracy from the North nearly bankrupted the colonial treasury. Furthermore, the state of near constant war caused a high death and desertion rate among the Mestizo, Mulatto and Indio (Native American) soldiers sent from Mexico and Peru that were stationed in the Philippines. The high death and desertion rate also applied to the native Filipino warriors conscripted by Spain, to fight in battles all across the archipelago. The repeated wars, lack of wages and near starvation were so intense, almost half of the soldiers sent from Latin America either died or fled to the countryside to live as vagabonds among the rebellious natives or escaped enslaved Indians (From India) where they race-mixed through rape or prostitution, further blurring the racial caste system Spain tried hard to maintain. These circumstances contributed to the increasing difficulty of governing the Philippines. The Royal Fiscal of Manila wrote a letter to King Charles III of Spain in which he advises to abandon the colony, but the religious orders opposed this since they considered the Philippines a launching pad for the conversion of the Far East.
Britain declared war against Spain on January 4, 1762 and on September 24, 1762 a force of British Army regulars and British East India Company soldiers, supported by the ships and men of the East Indies Squadron of the British Royal Navy, sailed into Manila Bay from Madras, India. Manila was besieged and fell to the British on October 4, 1762. The occupation of Manila ended in April 1764 as agreed to in the peace negotiations for the Seven Years' War in Europe. The Spanish then persecuted the Binondo Chinese community for its role in aiding the British. An unknown number of Indian soldiers known as sepoys, who came with the British, deserted and settled in nearby Cainta, Rizal, which explains the uniquely Indian features of generations of Cainta residents.
Spain and the United States had sent commissioners to Paris to draw up the terms of the Treaty of Paris to end the Spanish–American War. The Filipino representative, Felipe Agoncillo, had been excluded from sessions as Aguinaldo's government was not recognized by the family of nations. Although there was substantial domestic opposition, the United States decided to annex the Philippines. In addition to Guam and Puerto Rico, Spain was forced in the negotiations to cede the Philippines to the U.S. in exchange for US$20,000,000.00. U.S. President McKinley justified the annexation of the Philippines by saying that it was "a gift from the gods" and that since "they were unfit for self-government, ... there was nothing left for us to do but to take them all, and to educate the Filipinos, and uplift and civilize and Christianize them", in spite of the Philippines having been already Christianized by the Spanish over the course of several centuries. The First Philippine Republic resisted the U.S. occupation, resulting in the Philippine–American War (1899–1913)
Hostilities broke out on February 4, 1899, after two American privates killed three Filipino soldiers as American forces launched a major attack in San Juan, a Manila suburb. This began the Philippine–American War, which would cost far more money and take far more lives than the Spanish–American War.[172] Some 126,000 American soldiers would be committed to the conflict; 4,234 Americans died, as did 12,000–20,000 Philippine Republican Army soldiers who were part of a nationwide guerrilla movement of at least 80,000 to 100,000 soldiers.
The Negros Republic, formed in the Visayas under Aniceto Lacson prior to the formation of the First Philippine Republic, welcomed the advancing American army as a friendly force. Two other insurgent republics were briefly formed during American administration: the Tagalog Republic in Luzon, under Macario Sakay, and the Republic of Zamboanga in Mindanao under Mariano Arquiza.
When Democrat Woodrow Wilson became U.S. president in 1913, new policies were launched designed to gradually lead to Philippine independence. In 1902 U.S. law established Filipinos citizenship in the Philippine Islands; unlike Hawaii in 1898 and Puerto Rico in 1918, they did not become citizens of the United States. The Jones Law of 1916 became the new basic law, promised eventual independence. It provide for the election of both houses of the legislature. The Japanese invaded and occupied in 1942.
(The Philippines was named in honor of King Philip II of Spain).
 |
 |
 |
(Modern Vietnamese are admixed Black/Chinese and some French Mulattoes)
The ancient Chams/Champa of Vietnam present as Y-haplogroups
C-M216, C-M217 and C-M216*, K-P131*, R-M17, R-M124, and H-M69, O-M95*.
 |
 |
 |
The Cham people in Cambodia and Vietnam descend from refugees of the Kingdom of Champa, which once ruled much of Vietnam between Gao Ha in the north and Bien Hao in the south. A people of Malayo-Polynesian stock, the Cham developed under both Hindu and Muslim influence in their early history. The imprint of these two civilizations, although altered by local tradition and superstition, is still evident in the customs, mores, and religious practices of the Cham. Cham adherents of Hinduism and of Islam call themselves Cham Kaphir and Cham Bani respectively. The Vietnamese have historically considered the Cham culturally inferior, backward, and lazy. The Cham themselves prefer to remain separate from the Vietnamese; they strongly believe that only through isolation can they retain their cultural identity.
The Chams (also known as the Cham, Chiem Thanh, and Hroi in Vietnam) are a Malay people and the remnants of the ancient kingdom of Champa, which ruled southern Vietnam and Cambodia for more than 1000 years. They speak a Malay-Polynesian (Austronesian) language, similar to Indonesian, with Khmer, Vietnamese, Sanskrit, Indonesian and Arabic influences. They live primarily in south-central Vietnam and the Tonle Sap and Chau Doc areas of Cambodia.
For centuries a race of warriors and pirates, the Cham defended their vast and prosperous Kingdom of Champa from numerous invasions. However, in 1471, the empire finally collapsed before Vietnamese invaders. Only the grandiose temples and sanctuaries, irrigation systems, sculpture, woven cloth, and jewelry remain as evidence of this once great civilization. The descendants of the once powerful Cham are scattered along the eastern coast of the Republic of Vietnam and near the Cambodian border. These people now eke out a living as artisans, farmers, and fishermen. The Cham live in small village settlements, grouped according to matrilineal kinship ties. Their language belongs to the Malayo-Polynesian family and is related to the Rhade, Jarai, and Raglai tongues. The Cham have traditionally been very religious and perform daily rituals to appease animistic spirits while also practicing Islam and Hinduism.
 |
 |
 |
The Dogrib are one division of the widespread population of the Dene or Athapaskan-speaking peoples who, by archaeological and linguistic evidence, first entered western Alaska from Siberia by way of the Bering land bridge that existed during late Pleistocene times - about 33% of the Dogrib people are haplogroup C.
 |
Tanana, Athabaskan-speaking North American Indian group that lived along the headwaters of the Tanana River in what is now central Alaska. Traditionally, they were nomadic hunters, relying chiefly on caribou, moose, and mountain sheep for food and clothing - 42% of the Tanana people are haplogroup C.
 |
Cook Islands Māori are a Polynesian ethnicity originating in the Cook Islands. The Cook Islands is a self-governing country in free association with New Zealand and is part of the Realm of New Zealand. As such, Cook Islanders are New Zealand citizens. Well over 90 percent of Cook Islanders are either of full or partial descent of the native Polynesian people of the islands, who are known as Cook Islands Māori. Cook Islands Māori share many ancestral links with the Māori of New Zealand and the native people (Mā'ohi) of French Polynesia. Some Cook Islanders are also of other Polynesian, European (Papa'a), or Asian descent. 83% of Cook Islanders are haplogroup C2.
 |
 |
Flores (Indonesian: Pulau Flores) is one of the Lesser Sunda Islands, a group of islands in the eastern half of Indonesia - 39% of the Flores are haplogroup C2.
 |
 |
 |
Most current Indonesians are Mulattos. There is evidence of Arab Muslim traders entering Indonesia as early as the 8th century A.D. However, it was not until the end of the 13th century that the spread of Islam began.The Chinese arrived in the early 15th century. The Portuguese came in 1512, Dutch and British traders followed in 1602: Indians were later brought to Indonesia by the Dutch in the 19th century as indentured labourers to work on plantations located around Medan in Sumatra. While the majority of these came from South India, a significant number also came from the north. The Medan Indians included Hindus, Muslims and Sikhs.The Japanese invaded and occupied during World War II.
 |
 |
 |
 |
The Changpa are a semi-nomadic Tibetan people found mainly in the Changtang in Ladakh and in Jammu and Kashmir. A smaller number resides in the western regions of the Tibet Autonomous Region and were partially relocated for the establishment of the Changtang Nature Reserve. As of 1989 there were half a million nomads living in the Changtang area.
The homeland of the Changpa is a high altitude plateau known as the Changtang, which forms a portion of western and northern Tibet extending into southeastern Ladakh, and Changpa means "northerners" in Tibetan. Unlike many other nomadic groups in Tibet, the Changpa are not under pressure from settled farmers as the vast majority of land they inhabit is too inhospitable for farming.
Background: The Jomon and the Ainu were the original Black settlers of Japan. In 350 B.C. a Mongol group called the "Yayoi" break-off from China and invade, conquer and destroy their civilizations. These Yayoi are the progenitors of modern Japanese. The Jomon are extinct, like with Hawaiians, pure-blood Ainu no longer exist either.
The Ainu people are most closely related to the Jomon people. Ainu belong mainly to Y-haplogroup D-M55 (D1a2) and C-M217. A recent DNA study in 2019 suggests that haplogroup D-M55 was carried by about 70% and haplogroup C1a1 by about 30% of the ancient Jōmon people.
 |
 |
Haplogroup D (CTS3946)
Haplogroup D1 (M174) Found in Japan, China (especially Tibet), the Andaman Islands: Haplogroup D1a (CTS11577): Haplogroup D1a1 (Z27276, Z27283, Z29263): Haplogroup D1a1a (M15) Found mainly in Tibetans, Qiangic peoples, Yi, and Hmong-Mien peoples: Haplogroup D1a1b (P99) Found mainly in Tibetans, Qiangic peoples, Naxi, and Turkic peoples: Haplogroup D1a2 (M55, M57, M64.1, M179, P12, P37.1, P41.1 (M359.1), 12f2.2) Found mainly in Japan: Haplogroup D1a3 (Y34637) Found in Andamanese peoples (Onge, Jarawa): Haplogroup D1b (L1366, L1378, M226.2) Found in Mactan Island, Philippines: Haplogroup D2 (A5580.2) Found in Nigeria, Saudi Arabia and Syria.
ISOGG
Y-DNA haplogroup D is seen primarily in Central Asia, Southeast Asia, and in Japan and was established approximately 50,000 years ago. The high frequency of haplogroup D in Tibet (about 50%) and in Japan (about 35%) implies some early migratory connection between these areas. Examination of the genetic diversity seen in subgroup D1b-M64.1 in Japan implies that this group has been isolated in Japan for between 12,000-20,000 years. The highest frequencies of D1b-M64.1 in Japan are seen among the Ainu and the Ryukyuans.
 |
 |
E-M81 is found at an average frequency of 45% in the Maghreb and Libya, with peaks at over 60% in Tunisia as well as central and southern Morocco. It is especially common among Berber populations all over Northwest Africa, including the Tuaregs. Frequencies of over 75% have been reported among the Tuaregs of Burkina Faso and Mali.
 |
 |
In Europe, M81 is most common in Portugal (8%), Spain (4%), as well as in France (0-6%) and Italy (0-4%), where strong regional variations are observed. M81 is especially common in western Iberia, notably Extremadura (15.5%), Andalusia (13.5%), southern Portugal (11%), the Canary Islands (11%), north-west Castille (10%) and Galicia (10%). The highest percentage of E-M81 in Europe is found among the Pasiegos (30%, n=101), an isolated community living in the mountains of Cantabria.
 |
The highest frequencies of E-M123 are observed in Jordan (31% near the Dead Sea), Ethiopia (5-20%), Israel/Palestine (10-12% among the Palestinians and the Jews), among the Bedouins (8%), in Lebanon (5%), in North Africa (3-5%), Anatolia (3-6%) and southern Europe, particularly Italy (1 to 8%), in the Spanish region of Extremadura (4%), and the Balearic islands of Ibiza and Minorca (average 10%).
 |
It has been calculated that E-V13 emerged from E-M78 some 7,800 years ago, when Neolithic farmers were advancing into the Balkans and the Danubian basin. Furthermore, all the modern members of E-V13 descend from a common ancestor who lived approximately 5,500 years ago, and all of them also descend from a later common ancestor who carried the CTS5856 mutation. That ancestor would have lived about 4,100 years ago, during the Bronze Age. Almost immediately afterwards, CTS5856 split into six subclades, then branched off into even more subclades in the space of a few generations. In just a few centuries, that very minor E-V13 lineage had started an expansion process that would turn it into one of Europe's most widespread paternal lineages and reach far beyond the borders of Europe itself, also spreading to the eastern edge of the Mediterranean, the Caucasus, Kurdistan, Iran, and even Siberia.
This data suggests that the fate of E-V13 was linked to the elite dominance of Bronze Age society. The geographic distribution of the six main branches show that E-V13 quickly spread to all parts of Europe, but was especially common in Central Europe. The only Bronze Age migration that could account for such a fast and far-reaching dispersal is that of the Proto-Indo-Europeans. At present the most consistent explanation is that E-V13 developed from E-M78 in Central or Eastern Europe during the Neolithic period, and was assimilated by the R1a and R1b Proto-Indo-Europeans around the time that they were leaving the Pontic Steppe to invade the rest of Europe. (Typical Albino bullshit trying to give themselves a part in Human history that they really did NOT have.
 |
Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations |
The reason why we had to repeat our refrain, is because of the statement in pink (or magenta if you like) above.
The Albinos are liars yes: but they aren't stupid, they know perfectly well that any decent Encyclopedia will detail
how Albinos starting with the Germanic tribes at the dawn of the MODERN era, then the Turks in the middle ages,
Then the French and Italians in our time, colonized North Africa and produced all of the Albinos and their Mulattoes
that you find running around the place today.
Wiki:
Taforalt or Grotte des Pigeons, is a cave in northern Oujda, Morocco, and possibly the oldest cemetery in North Africa (Humphrey et al. 2012). It contained at least 34 Iberomaurusian (pertaining to a population of early Humans that occupied the Mediterranean littoral from Morocco to Tunisia 22,000 to 9,000 years ago) adolescent and adult human skeletons, as well as younger ones, from the Upper Palaeolithic between 15,100 and 14,000 calendar years ago. There is archaeological evidence for Iberomaurusian occupation at the site between 23,200 and 12,600 calendar years ago, as well as evidence for Aterian occupation as old as 85,000 years.
Ancient DNA
In 2018, van de Loosdrecht et al. performed the first DNA tests on the ancient Taforalt individuals, directly dated to between 15,100 and 13,900 ya. The Taforalt samples are the oldest human DNA samples from Africa yet recovered. DNA analysis was performed on seven individuals: six males and one female. Only five of individuals, including four of the males, with higher coverage genomes were used in the nuclear DNA analysis. Nuclear DNA analysis reveals that the Taforalt individuals were all closely related to each other, showing evidence of a population bottleneck event in their past."
The Taforalt genomes were found to be composed of three major components: a Holocene Levantine component, a Hadza hunter-gatherer component from Tanzania (Genetically, the Hadza are not closely related to any other people), and a West African component. The Taforalt individuals show closest genetic affinity for ancient Epipaleolithic Natufian individuals (Mesolithic culture of ancient Palestine characterized by microliths), with slightly better affinity for the Natufians than later Neolithic Levantines. A two-way admixture scenario using Natufian and modern West African samples as reference populations inferred that the Taforalt individuals bore 63.5% Natufian-related and 36.5% West African-related ancestries, with no evidence for additional gene flow from the Epigravettian culture of Upper Paleolithic Europe. The Taforalt individuals also show evidence of limited Neanderthal ancestry.
When compared against modern populations, the Taforalt individuals form a distinct cluster and do not cluster genetically with any modern population; however, they were found to cluster between modern North Africans and East Africans. The Taforalt individuals also exhibit higher levels of Sub-Saharan and Hadza-related ancestry than do modern North Africans.
 |
 |
 |
 |
 |
 |
 |
 |
|
His Royal Highness King Mohammed VI of Morocco |
Her Royal Highness Princess Lalla Salma of Morocco |
Outside Europe, E1b1b (formerly known as E3b) is found at high frequencies in Morocco (over 80%), Somalia (80%), Ethiopia (40% to 80%), Tunisia (70%), Algeria (60%), Egypt (40%), Jordan (25%), Palestine (20%), and Lebanon (17.5%). On the European continent it has the highest concentration in Kosovo (over 45%), Albania and Montenegro (both 27%), Bulgaria (23%), Macedonia and Greece (both 21%), Cyprus (20%), Sicily (20%), South Italy (18.5%), Serbia (18%) and Romania (15%). Ashkenazi Jews have approximately 20% of E1b1b, which falls mostly under specific clades of E-M123. E-M81 is the most common subclade of haplogroup E-L19/V257. It is concentrated in the Maghreb and is dominated by its E-M183 subclade.
E-M183 is believed to have originated in northwestern Africa. Typical of E-M183 people are the Reguibat Nomads of the Sahrawi tribe, which are of of Sanhaja-Berber origins. The Reguibat speak Hassaniya Arabic, and are Arabized in culture. They claim descent from Sidi Ahmed Rguibi, who lived in the Saguia el-Hamra region in the 16th century. They also believe that they are, through him, a chorfa tribe, i.e. descendants of Muhammad. Religiously, they belong to the Maliki school of Sunni Islam.
Initially they were an important Arabic zawiya or religious tribe with a semi-sedentary lifestyle, but the Reguibat gradually turned to camel-rearing, raiding and nomadism during the 18th century. In response to attacks from neighboring tribes which provoked them into taking up arms and leaving the subordinate position they had previously held. This started a process of rapid expansion, and set the Reguibat on the course towards total transformation into a traditional warrior tribe in the late 19th century. By then they had become well-established as the largest Sahrawi tribe, and were recognized as the most powerful warrior tribe of the area.
 |
The grazing lands of the Reguibat fractions extended from Western Sahara into the northern half of Mauritania, the edges of southern Morocco and northern Mali, and large swaths of western Algeria (where they captured the town of Tindouf from the Tajakant tribe in 1895, and turned into an important Reguibat encampment). The Reguibat were known for their skill as warriors, as well as for an uncompromising tribal independence, and dominated large areas of the Sahara desert through both trade and use of arms.
Reguibat Sahrawis were very prominent in the resistance to French and Spanish colonization in the 19th and 20th century, and could not be subdued in the Spanish Sahara until 1934, almost 50 years after the area was first colonized by Spain. Since the 1970s, many Reguibat have been active in the Polisario Front's resistance to Moroccan rule over the still non-sovereign Western Sahara territory. Polisario leader Mohamed Abdelaziz was Reguibi, as is the Moroccan CORCAS leader Khalihenna Ould Errachid.
Haplogroup E-M96 is a human Y-chromosome DNA haplogroup. It is one of the two main branches of the older and ancestral haplogroup DE, the other main branch being haplogroup D. The E-M96 clade is divided into two main subclades: the more common E-P147, and the less common E-M75.
Origins
Underhill (2001) proposed that haplogroup E may have arisen in East Africa. Some authors as Chandrasekar (2007), accept the earlier position of Hammer (1997) that Haplogroup E may have originated in Asia, given that:
Underhill and Kivisild (2007) demonstrated that C and F have a common ancestor meaning that DE has only one sibling which is non-African.
DE* is found in both Asia and Africa, meaning that not only one, but several siblings of D are found in Asia and Africa.
Karafet (2008), in which Hammer is a co-author, significantly rearranged time estimates leading to "new interpretations on the geographical origin of ancient sub-clades". Amongst other things this article proposed a much older age for haplogroup E-M96 than had been considered previously, giving it a similar age to Haplogroup D, and DE itself, meaning that there is no longer any strong reason to see it as an offshoot of DE which must have happened long after DE came into existence and had entered Asia.
Y- HAPLOGROUP E1b1 (E-P2)
E1b1 (E-P2) the most dominant Y-chromosome haploid in Africa. E-P2 is likely to have originated in the Ethiopian highlands of East Africa, as this is the place with the high frequency of ancestral sub-clades of this haplogroup. E-P2 diverged into two predominant sub-clades; E1b1a (E-V38) and E1b1b (E-M215) approximately 24-27,000 years ago.
Haplogroup E1b1a
Haplogroup E1b1a is the main haplogroup in sub-Saharan Africa. Over 80% of the males in West Africa fall under this haplogroup. and others suggest that it likely originated in and expanded from West Africa (i.e., the Sudan Belt) during the last 20,000 to 30,000 years based on the fact that the frequency and divergence of E1b1a in this region are notably the highest found.
With respect to E1b1a, a west-to-east as well as a south-to-north clonal distribution exists, that is, the diversity and frequency increases as you move from East and North Africa to West and South of Africa. That’s why it is observed in low frequencies in the Horn of Africa, North Eastern Africa, and Southwest Asia, where the E1b1b haplogroup has its lowest frequencies, and its meager presence in these areas is generally characteristic to the slave commerce and/or the Bantu expansion through past migrations.
E1b1a Sub-clades
E1b1a is the single most common Y-chromosome haplogroup among people of Sub-Saharan African descent both inside and outside of Africa. It has been observed at frequencies of 58%-60% of African American populations. The E1b1a sub clades E1b1a7 and E1b1a8 are widely found throughout sub-Saharan Africans. However, the sub-clade E1b1a9 has been found only in one Gambian and also the sub-clades E1b1a2, E1b1a3, E1b1a4, E1b1a5, and E1b1a6 are quite rare as well.
2nd. King of the 20th. Dynasty
Objective To investigate the true character of the harem conspiracy described in the Judicial Papyrus of Turin and determine whether Ramesses III was indeed killed. Design Anthropological, forensic, radiological, and genetic study of the mummies of Ramesses III and unknown man E, found together and taken from the 20th dynasty of ancient Egypt (circa 1190-1070 BC).
Results Computed tomography scans revealed a deep cut in Ramesses III’s throat, probably made by a sharp knife. During the mummification process, a Horus eye amulet was inserted in the wound for healing purposes, and the neck was covered by a collar of thick linen layers. Forensic examination of unknown man E showed compressed skin folds around his neck and a thoracic inflation. Unknown man E also had an unusual mummification procedure. According to genetic analyses, both mummies had identical haplotypes of the Y chromosome and a common male lineage.
Conclusions This study suggests that Ramesses III was murdered during the harem conspiracy by the cutting of his throat. Unknown man E is a possible candidate as Ramesses III’s son Pentawere. Ramesses III Y-dna haplogroup is E1b1a.
Haplogroup E1b1b
E1b1b is found at high frequencies outside Europe, over 80% in Morocco and East Africa Somalia and Ethiopia males makes about 40% to 80%. North African countries of Tunisia, (70%), Algeria (60%) and Egypt (40%). In the Middle East region; Jordan (25%), Palestine (20%), and Lebanon (17.5%). On the European continent, Eastern and Central Europe, Kosovo has the highest concentration over 45%, Albania and Montenegro (both 27%), Bulgaria (23%), Macedonia and Greece (both 21%), the Island of Cyprus and Sicily (both 20%), South Italy (18.5%), Serbia (18%) and Romania (15%).
Haplogroup E1b1b (formerly known as E3b) represents the last major direct exodus from Africa into Europe believed to have appeared first in the Horn of Africa about 26,000 years ago and scattered to North Africa and the Near East during the late Paleolithic and Mesolithic periods. E1b1b lineages are closely linked to the diffusion of Afro-asiatic languages.
The highest genetic diversity of haplogroup E1b1b is noted in Northeast Africa region in Ethiopia and Somalia, which also have the monopoly of older and rarer sub-clades like M281, V6 or V92.
More specifically, Ethiopia and Somali males belong mostly to the V22 and V32 sub-clades, but possess also a minority of M81, M123 and V42 sub clades.
E1b1b Sub-clades
 |
Haplogroup E (M96)
Haplogroup E (M40, M96) Found in Africa and parts of the Middle East and Europe: Haplogroup E1 (P147): Haplogroup E1a (M33, M132) formerly E1: Haplogroup E1b (P177): Haplogroup E1b1 (P2, DYS391p); formerly E3: Haplogroup E1b1a (V38): Haplogroup E1b1a1 (M2) Found in Africa, especially among Niger–Congo-speaking populations.; formerly E3a: Haplogroup E1b1a2 (M329) Found in Africa, especially in Ethiopia among Omotic-speaking populations.; formerly E3*: Haplogroup E1b1b (M215): Haplogroup E1b1b1 (M35) Found in Horn of Africa, North Africa, the Middle East, and Europe (especially in areas near the Mediterranean and the Balkans); formerly E3b: Haplogroup E2 (M75).
ISOGG
Y-DNA haplogroup E would appear to have arisen in northern hemisphere of Africa based on the concentration and variety of E subclades in that area today. E1b1 is by far the lineage of greatest geographical distribution. It has two major sub-lineages, E1b1a and E1b1b. E1b1a is a lineage that originated and expanded from West or Central Africa to Eastern Africa and Southern Africa. E1b1b probably evolved either in Northeast Africa or the Near East and then expanded to the west--both north and south of the Mediterranean Sea. E1b1b1 clusters are seen today in Western Europe, Southeast Europe, the Near East, Northeast Africa and Northwest Africa. E2 is possibly of Central African origin and is spread in low frequencies across the continent. The Cruciani articles (references and links below) are indispensable resources for understanding the structure of this complicated haplogroup, but note that the Cruciani haplogroup labels are now superseded because of the recently discovered new SNPS that lie closer to the root of the E branch of the Y-haplogroup Tree.
 |
Haplogroup F, also known as F-M89 and previously as Haplogroup FT is a very common Y-chromosome haplogroup. The clade and its subclades constitute over 90% of paternal lineages outside of Africa. It is primarily found throughout South Asia, Southeast Asia and parts of East Asia.
The vast majority of individual males with F-M89 fall into its direct descendant Haplogroup GHIJK (F1329/M3658/PF2622/YSC0001299). In addition to GHIJK, haplogroup F has three other immediate descendant subclades: F1 (P91/P104), F2 (M427/M428), and F3 (M481). These three, with F* (M89*), constitute the paragroup F(xGHIJK).
Haplogroup GHIJK branches subsequently into two direct descendants: G (M201/PF2957) and HIJK (F929/M578/PF3494/S6397). HIJK in turn splits into H (L901/M2939) and IJK (F-L15). The descendants of the haplogroup IJK include the clades I, J, K, and, ultimately, several major haplogroups descended from Haplogroup K, namely: haplogroups M, N, O, P, Q, R, S, L, and T.
(ISOGG) International Society of Genetic Genealogy - 2018.
Y-DNA haplogroup F is the parent of all Y-DNA haplogroups G through T and contains more than 90 percent of the world's population. Haplogroup F was in the original migration out of Africa, or else it was founded soon afterward, because F and its sub-haplogroups are primarily found outside, with very few inside, sub-Saharan Africa. The founder of F could have lived between 60,000 and 80,000 years ago, depending on the time of the out-of-Africa migration.
The major sub-groups of Haplogroup F are Haplogroups G, H, [IJ], and K, which are discussed elsewhere at this site. The minor sub-groups have not been well studied, but apparently occur only infrequently and primarily in southern parts of Asia.
Haplogroup F (M89)
A hypothetical diversion of Haplogroup F and its descendants. The groups descending from haplogroup F are found in some 90% of the world's population, but almost exclusively outside of sub-Saharan Africa. F xG,H,I,J,K is rare in modern populations and peaks in South Asia, especially Sri Lanka. It also appears to have long been present in South East Asia; it has been reported at rates of 4–5% in Sulawesi and Lembata. One study, which did not comprehensively screen for other subclades of F-M89 (including some subclades of GHIJK), found that Indonesian men with the SNP P14/PF2704 (which is equivalent to M89), comprise 1.8% of men in West Timor, 1.5% of Flores 5.4% of Lembata 2.3% of Sulawesi and 0.2% in Sumatra. F* (F xF1,F2,F3) has been reported among 10% of males in Sri Lanka and South India, 5% in Pakistan, as well as lower levels among the Tamang people (Nepal), and in Iran. F1 (P91), F2 (M427) and F3 (M481; previously F5) are all highly rare and virtually exclusive to regions/ethnic minorities in Sri Lanka, India, Nepal, South China, Thailand, Burma, and Vietnam. In such cases, however, the possibility of misidentification is considered to be relatively high and some may belong to misidentified subclades of Haplogroup GHIJK.
ISOGG
"Geographical distribution so far of haplogroup F subgroups (not complete): F1 - P91 Sri Lanka: F2 - M427 China (Lahu, Yi), s.e. Asia: F3 - M481 s.e. India: F4 - Z40733 Vietnam".
 |
 |
 |
 |
Haplogroup G (M201)
Haplogroup G (M201) originated some 48,000 years ago and its most recent common ancestor likely lived 26,000 years ago in the Middle East. It spread to Europe with the Neolithic Revolution. It is found in many ethnic groups in Eurasia; most common in the Caucasus, Iran, Anatolia and the Levant. Found in almost all European countries, but most common in Gagauzia, southeastern Romania, Greece, Italy, Spain, Portugal, Tyrol, and Bohemia with highest concentrations on some Mediterranean islands; uncommon in Northern Europe. G-M201 is also found in small numbers in northwestern China and India, Bangladesh, Pakistan, Sri Lanka, Malaysia, and North Africa.
ISOGG
Researchers have also suggested various places in western Asia as the site of origin of G, but the lack of ancient DNA from that period makes confirmation of this difficult. A unitary concept of haplogroup G often has little practical importance because virtually all G men belong to G subgroups that arose much more recently and have differing geographical distributions.
| Quote: We know from archeological data that in the upper Paleolithic period Anatolia was settled by populations with Aurignacian culture (Kuhn 2002). Recent genetic studies (Cinnioglu et al. 2004; Olivieri et al. 2006) based on the analysis of mtDNA (haplogroup M1 and U6) and the Y chromosome (R1b3-M269 lineage) suggest, in agreement with paleoenvironmental evidence (van Andel and Tzedakis 1996), that around 40,000–45,000 years ago, populations with Aurignacian culture may have spread by migration from the Levant and southwest Asia to Anatolia and further into Europe (Bar-Yosef 2002). With the exception of these scarce molecular data, almost nothing is known about the biological features of these early Paleolithic Anatolian foragers. Nevertheless, considering the important demographic processes and biological changes undergone by human populations as a result of later and major events (e.g., the Neolithic transition), we believe that the causes of the observed affinity patterns have to be determined from these later periods.
From the Mesolithic to the early Neolithic period different lines of evidence support an out-of-Africa Mesolithic migration to the Levant by northeastern African groups that had biological affinities with sub-Saharan populations.
{The Levant = Cyprus, Israel, Iraq, Jordan, Lebanon, Palestine, Syria, southern Turkey}. |
Native Anatolian (now called Turkey) |
 |
Research has been conducted to study the genetic origins of the modern Turkish people (not to be confused with the ethnic Turkic peoples originating in Asia) in Turkey. These studies sought to determine whether the modern Turks have a stronger genetic affinity with the Turkic peoples of Central Asia where the Seljuk Turks began their migrating to Anatolia. This migration led to the establishment of the Anatolian Seljuk Sultanate in the late 11th century. Or whether modern Turkish people instead largely descended from the indigenous peoples of Anatolia who were culturally assimilated during the Seljuk and Ottoman periods.
Of course we all remember when Albinos once claimed that Haplogroup "R" was an indicator of "White" people. Now we know that the only indicator of White people is one of the seven "Mutated" genes which cause Albinism. See one of the Albinism tables on this and other pages. To see what "Pure" (R) people look like, see the haplogroup (R) section below. |
 |
 |
|
Altai Turks in Russian Siberia |
Altai Turks in Russian Siberia |
 |
(Turks defeated the Black Eastern Roman Empire (the Byzantine) in 1453 A.D.)
 |
 |
 |
 |
Haplogroup H (M69)
Haplogroup H (M69) probably emerged in South Asia, about 48,000 years BP, and remains prevalent there, in the forms of H1 (M69) and H3 (Z5857). However, H2 (P96) has been present in Europe since the Neolithic and H1a1 (M82) spread westward in the Medieval era with the migration of the Romani.
H-M69 is common among populations of India, Sri Lanka, and Nepal, with lower frequency in Afghanistan and Pakistan. ... and H-M52 among Kalash (20.5%) in Pakistan. Haplogroup H is typically found among Dravidian populations in the Indian subcontinent, especially in South India and Sri Lanka.
 |
 |
 |
 |
 |
An Albino anthropologist greatest joy is to stealthily insert a bit of data that makes it seem like Central Asian Albinos (modern White people) were in Europe from earlier than modern times: except of course for the first incursion, circa 1,200 B.C. Note the number of Albino scientist who signed on to the subtle lying of the study below: "The Beaker Phenomenon and the Genomic Transformation of Northwest Europe". It's about 150 co-liars (authors), more than we had seen before on any study. But as you can see, these liar scientists, all 150 of them, were beaten to the punch by truth - see below. For facts on the Beaker Culture, please see the Etruria-3 section.
The Bell Beaker cultureThe Bell Beaker culture (or, in short, Beaker culture) is an archaeological culture named after the inverted-bell beaker drinking vessel used at the very beginning of the European Bronze Age. Arising from around 2,800 BC, it lasted in Britain until as late as 1800 B.C, but in continental Europe only until 2,300 B.C, when it was succeeded by the Unetice culture. The culture was widely dispersed throughout Western Europe, from various regions in Iberia (Spain and Portugal) and spots facing northern Africa to the Danubian plains, the islands of Great Britain and Ireland, and also the islands of Sicily and Sardinia. |
Haplogroup I (M170)
Haplogroup I (M170, M258) is found mainly in Europe and the Caucasus. Haplogroup I1 (M253) Found mainly in northern Europe: Haplogroup I2 (P215) Found mainly in Balkans, southeast Europe and Sardinia save for I2B1 (m223) which is found at a moderate frequency in Western, Central, and Northern Europe.
ISOGG
Y-DNA haplogroup I is an (original Black European) haplogroup, representing nearly one-fifth of the population. It is almost non-existent outside of Europe, suggesting that it arose in Europe. Estimates of the age of haplogroup I suggest that it arose prior to the last Glacial Maximum, long before Whites/Albinos entered Europe.
The two main subgroups of haplogroup I likely divided approximately 28,000 years ago.
I1-M253 et al has highest frequency in Scandinavia, Iceland, and northwest Europe. In Britain, haplogroup I-M253 et al is often used as a marker for "invaders," Viking or Anglo-Saxon. The I-M227 subclade is concentrated in eastern Europe and the Balkans and appears to have arisen in the last one thousand to five thousand years. It has been reported in Germany, Czech Republic, Poland, Estonia, Ukraine, Switzerland, Slovenia, Bosnia, Macedonia, Croatia, and Lebanon.
I2-M438 et al includes I2* which shows some membership from Armenia, Georgia and Turkey; I-P37.2, which is the most common form in the Balkans and Sardinia. I-M26 is especially prevalent in Sardinia. I2a2-M436 et al reaches its highest frequency along the northwest coast of continental Europe. I-M223 et al occurs in Britain and northwest continental Europe. I2a2a1-M284 occurs almost exclusively in Britain, so it apparently originated there and has probably been present for thousands of years.
 |
 |
 |
 |
| Today 26% of the English are Y-Haplogroup (I): Dutch 33%, Germans 24%, Abkhazians 33%, Austrians 28%, Belarusians 32%, Bosnia and Herzegovina 65%, Bulgarians 29%, Croats 46%, Danes 39%, Darginians 58%, Flemish & Belgians 28%, Finland 29%, Hungarians 28%, Macedonia 24%, Norwegians 45%, Romanians 28%, Saami 31%, Serbs 42%, Slovenians 30%, Swedes 44%, Ukrainians 22%, |
 |
Bosnia and Herzegovina |
 |
Darginians (Dagestan) |
 |
Norway |
 |
Swedes |
 |
Haplogroup J (M304)
Haplogroup J (M304, S6, S34, S35) is found mainly in the Middle East and South-East Europe. Haplogroup J* (J-M304*) is rare outside the island of Socotra. Haplogroup J1 (M267) is associated with Northeast Caucasian peoples in Dagestan and Semitic languages speaking people in the Middle East, Ethiopia, and North Africa and also found in Mediterranean Europe in smaller frequencies much like haplogroup T: Haplogroup J2 (M172) is found mainly in the Semitic-speaking peoples, Anatolia, Greece, the Balkans, Italy, Iran, the Caucasus, South Asia, and Central Asia.
A majority of Chechens belong to Haplogroup J2 (56.7%), which is associated with Mediterranean, South Caucasian and Fertile Crescent populations, with its peaks at 87.4% in Ingushetia and 72% in Georgia's Kazbegi Municipality.
ISOGG
"Y-DNA haplogroup J evolved in the ancient Near East and was carried into North Africa, Europe, Central Asia, Pakistan and India. J2 lineages originated in the area known as the Fertile Crescent. The main spread of J2 into the Mediterranean area is thought to have coincided with the expansion of agricultural peoples during the Neolithic period. The timing of the demographic events that brought J2 to Central Asia, Pakistan, and India is not yet known. J1 lineages may have a more southern origin, as they are more often found in the Levant region, other parts of the Near East, and North Africa, with a sparse distribution in the southern Mediterranean flank of Europe, and in Ethiopia.
"There is a descending gradient in the frequency of occurrence of haplogroup J from the Middle East toward the northwest of Europe, reaching about 3% of the population on the northwest Atlantic coast. The occurrence of J in Europe is undoubtedly due both to the Neolithic expansion and to episodic migrations, though the relative proportion of those two sources is controversial and may not be the same in different locations. A significant fraction of Jews belong to haplogroup J, but Jews represent a small minority of the European members of the haplogroup. The ""Cohen Modal Haplotype"" is a specific set of six Y-STR marker values that occurs in both J1 and J2, though at a much higher frequency in J1.
King Peroz I;
This king was independently identified as Peroz I: he was the eighteenth king of the
Sassanian Empire from 459 A.D. to 484 A.D. He was the son of Yazdegerd II
and younger brother of Hormizd III, whom Peroz I seized the throne from.
Abstract
Knowledge of high resolution Y-chromosome haplogroup diversification within Iran provides important geographic context regarding the spread and compartmentalization of male lineages in the Middle East and southwestern Asia. At present, the Iranian population is characterized by an extraordinary mix of different ethnic groups speaking a variety of Indo-Iranian, Semitic and Turkic languages. Despite these features, only few studies have investigated the multiethnic components of the Iranian gene pool. In this survey 938 Iranian male DNAs belonging to 15 ethnic groups from 14 Iranian provinces were analyzed for 84 Y-chromosome biallelic markers and 10 STRs. The results show an autochthonous but non-homogeneous ancient background mainly composed by J2a sub-clades with different external contributions. The phylogeography of the main haplogroups allowed identifying post-glacial and Neolithic expansions toward western Eurasia but also recent movements towards the Iranian region from western Eurasia (R1b-L23), {comment: Dravidian Albinos}, Central Asia (Q-M25), Asia Minor (J2a-M92) and southern Mesopotamia (J1-Page08).
In 2004 the National Geographic Genographic Project, Dr. Pierre Zalloua and Dr. Spencer Wells, identified "the haplogroup of the Phoenicians" as Y-haplogroup J2. The beginning of the end for the Phoenicians of the Canaanite City States, came when Cyrus the Great conquered Phoenicia in 539 B.C. The Persians divided Phoenicia into four vassal kingdoms: Sidon, Tyre, Arwad, and Byblos. The Phoenicians prospered furnishing fleets for the Persian kings, but Phoenician influence declined after this. In 350 or 345 B.C. a rebellion in Sidon led by Tennes (king of Sidon) was crushed by Persian king Artaxerxes III.
 |
 |
Carthage was founded on the north coast of Africa (Tunisia) by the Canaanites/Phoenicians of the city of Tyre in 814 B.C. The founding of Carthage was closely followed by the establishment of other Phoenician cities in the western Mediterranean. This brought them into direct conflict with the Empires in Rome and Greece. At the start of the 3rd century B.C., Carthage was supreme in the western Mediterranean, enjoying the security of sea power and trading with her stations in Sicily, Sardinia, and Spain as well as with the shores of Africa. At the time Rome was struggling to obtain mastery over central and southern Italy, where it had absorbed the power and culture of the Etruscans, and gradually forged a federation of small states. It must have already become clear that there was not going to be enough room in the Mediterranean for both Rome and Carthage.
The First Punic War (264–241 B.C.) was fought partly on land in Sicily and Africa, but was largely a naval war. It began as a local conflict in Sicily between Hiero II of Syracuse and the Mamertines of Messina. The Mamertines enlisted the aid of the Carthaginian navy, and subsequently betrayed them by entreating the Roman Senate for aid against Carthage. The Romans sent a garrison to secure Messina, so the outraged Carthaginians then lent aid to Syracuse. Tensions quickly escalated into a full-scale war between Carthage and Rome for the control of Sicily. After a harsh defeat at the Battle of Agrigentum in 262 B.C., the Carthaginian leadership resolved to avoid further direct land-based engagements with the powerful Roman legions, and concentrate on the sea where they believed Carthage's large navy had the advantage. Initially the Carthaginian navy prevailed, but eventually lost.
With Carthage paying reparations to Rome, Hamilcar Barca, had popular support and the command of the armed forces. With these he proceeded to increase Carthaginian hold on Spain; ostensibly to enable Carthage to pay repatriation to Rome, but in fact, because he saw in Spain as a source of manpower and supplies and a base from which to attack Rome. With his son-in-law Hasdrubal and his four sons Hannibal, Hasdrubal, Hanno, and Mago, the 'lion's brood' as he called them. Hamilcar barca soon succeeded in turning southern Spain into a sort of empire where a new Carthage or Carthagena was founded. In 228 B.C. he fell in battle and was succeeded by hasdrubal his son-in- law who, in his turn was murdered seven years later in 221 B.C. The army thereupon unanimously chose Hannibal to be their general in spite of his youth "because of the shrewdness and courage which he had shown in their service." Hannibal, whose name means "Joy of Baal" (Canaanite God), was then 26 years old, he accompanied his father on his campaign in Spain, at the tender age of nine.
The Second Punic War (218–201 B.C.) is remembered for Hannibal's crossing of the Alps. His army invaded Italy from the north and resoundingly defeated the Roman army in several battles, but never achieved the ultimate goal of causing a political break between Rome and its allies.
The Third Punic War (149–146 B.C.) involved an extended siege of Carthage, ending in the city's thorough destruction. In 149 B.C., in an attempt to draw Carthage into open conflict, Rome made a series of escalating demands, one being the surrender of three hundred children of the nobility as hostages, and finally ending with the near-impossible demand that the city be demolished and rebuilt away from the coast, deeper into Africa. When the Carthaginians refused this last demand, Rome declared the Third Punic War. A second offensive under the command of Scipio Aemilianus resulted in a three-year siege before he breached the walls, sacked the city, and systematically burned Carthage to the ground in 146 B.C. When the war ended, the remaining 50,000 Carthaginians, a small part of the original pre-war population, were sold into slavery. Carthage was systematically burned for 17 days; the city's walls and buildings were utterly destroyed. The remaining Carthaginian territories were annexed by Rome and reconstituted to become the Roman province of Africa.
When Carthage fell, its nearby rival, Utica Tunisia, (also Phoenician), a Roman ally, was made capital of the region and replaced Carthage as the leading center of Punic trade and leadership. It had the advantageous position of being situated on the outlet of the Medjerda River, Tunisia's only river that flowed all year long. However, grain cultivation in the Tunisian mountains caused large amounts of silt to erode into the river. This silt accumulated in the harbor until it became useless, and Rome was forced to rebuild Carthage.
Over 100 years later, a new city of “ROMAN” Carthage was built on the same land by Julius Caesar in the period from 49 to 44 B.C., and by the first century, it had grown to be the second-largest city in the western half of the Roman Empire, with a peak population of 500,000. It was the center of the province of Africa, which was a major breadbasket of the Empire. Among its major monuments was an amphitheater.
| Concerning the coin above: at first Albinos said that the image was not of Hannibal, but rather his "Mahout" (Elephant rider). As ridiculous as that sounds, it is not new for Albinos, they say the same kind of thing about the 1792 Barbados Penny. | |
| So lets flesh out this thinking for Americans: Lets say that after the war of independence is won, they wish to commemorate the triumph of a great leader like George Washington; so they create a coin or painting of him crossing the Delaware River. But in the bow of the canoe, instead of showing Washington, they show his SERVANT!!!! | |
| The above was just too stupid, even for the most ignorant to believe, so a new lie was needed. So to try and create confusion, some Albinos have taken to asserting that their neighbors, the Berbers, were Albinos. Example: Quote - Here is a coin of one of their allies, Masinissa, King of Numida (a Berber). | |
And right on time, the Albinos at Wikipedia come up with a fake coin showing an Albino Berber.
|
|
|
The Albinos have invested their entire racial identity in being Greeks and Romans, or rather, the descendants of Greeks and Romans. Most of their FAKE statuary are of Greeks and Romans. Which is really strange when you consider that they have known for quite some time that they are actually Central Asians who "CAME" into Europe: well actually they were "CHASED" into Europe by the Mongols.
 |
 |
We call the original inhabitants of the European Aegean area "Pelasgians", these people were no doubt akin to the Grimaldi people in the West. So far, the Albinos have not seen fit to do the Genome of the Grimaldi or Pelasgians. But no matter, the surrounding evidence assure us that they sure weren't Albinos.
The actual Greek "Civilization" is another matter, that we are sure of: it was established in Greece as the city of Mycenae circa 2,000 B.C. and it came from Crete, who got it's basic's from Egypt. According to the Scientific Study "Ancient DNA analysis reveals Minoan and Mycenaean origins" the major Y-dna haplogroups of those people were J2a1, J1a, G2a2b2.
Throughout the pages of Realhistoryww we have exposed countless examples of Albino Race lies. But just as damaging to seekers of truth as the told lie, is the untold truth. Here we see how these 32 degenerate authors of this study, conspired to hide the "Skin Color" of the Minoan's and Mycenaean's by duplicity appearing to gather data on their appearance while carefully avoiding the easiest data to gather, that is, data on their Skin Color via the Genes shown above.
Note that these pathetic Albinos looked for Blue and Brown eyes; Blonde, Brown, Red, and Black hair. And determined that as was normal with Blacks, their Eye and Hair color was mostly Brown and Black. But not a word about their skin color, because they knew the genes would show to be free of damage/Mutations, and thus Black genes: which is something Albinos are loath to admit.
But to be sure, we are not suggesting that the Albinos only arrived in Europe in the "Modern Era", there is circumstantial evidence (tomb paintings and the assumed reason for the "Sea People" exodus) which suggest that the first wave of Albinos arrived in Europe circa 1,200 B.C.
 |
 |
 |
The original civilization in Italy was the Etruscans, as with Greece, we are not sure where and how the terms the Albinos use, such as Latin's and Romans, come into play; (sometimes it's hard to determine what was totally made up by them as to who was, and did what). Likewise, no one seems to really know when the civilization started.
Main Y-haplogroup J2a-M67*
Mtdna below |
 |
 |
The South African Medical Journal
Lemba origins revisited: Tracing the ancestry of Y chromosomes in South African and Zimbabwean Lemba
The most common haplogroup in the Remba was J-M172, which accounted for 42.6% of their Y chromosomes, whereas J-12f2a was most common in the Lemba (39.5%). Both J haplogroups were observed in Jews at frequencies of 18.1% and 22.7%, respectively. The CMH (14-16-23-10-11-12), which was resolved on haplogroup J-12f2a was only found in the Lemba (13.2%) and Jewish (15.9%) groups, but not in the Remba (H54, Appendix 1). With the exception of L-M11, which was only found in Lemba and Remba, all haplogroups derived from non-African ancestry in the combined Lemba/Remba sample were also found in Jews.
This research published in 2013 in the South African Medical Journal studied Y-chromosomes variations in two groups of Lemba, one South African and the other Zimbabwean (the Remba). It concluded, "While it was not possible to trace unequivocally the origins of the non-African Y chromosomes in the Lemba and Remba, this study does not support the earlier claims of their Jewish genetic heritage." The researcher suggested "a stronger link with Middle Eastern populations, probably the result of trade activity in the Indian Ocean."
 |
 |
 |
Chechnya |
 |
ISOGG
Y-DNA haplogroup K
is an old lineage established approximately 40,000 thousand years ago whose origins were probably in southwestern Asia. K's structure is interwoven with other haplogroups downstream. The subclades restricted to K itself are found at low frequencies in various parts of Africa, Eurasia, Australia and the South Pacific. Geographical Distribution of ancestry of men in Haplogroup K (not complete): K2c P261 Balinese Indonesia: K2d P402 Javanese Indonesia: K2e M147 India, Pakistan.
Haplogroup K (M9)
Haplogroup K (M9) is spread all over Eurasia, Oceania and among Native Americans: K(xLT,K2a,K2b) – that is, K*, K2c, K2d or K2e – is found mainly in Melanesia, Aboriginal Australians, India, Polynesia and Island South East Asia.
Haplogroups K2b1, M & S
No examples of the basal paragroup K2b1* have been identified. Males carrying subclades of K2b1 are found primarily among Papuan peoples, Micronesian peoples, indigenous Australians, and Polynesians. Its primary subclades are two major haplogroups: Haplogroup S (B254) also known as K2b1a: found in the highlands of Papua New Guinea and; Haplogroup M (P256) also known as K2b1b: found in New Guinea and Melanesia.
'Ust'-Ishim man is the term given to the 45,000-year-old remains of one of the early modern humans to inhabit western Siberia. The fossil is notable in that it had intact DNA which permitted the complete sequencing of its genome, the oldest modern human genome to be so decoded.
The remains consist of a single bone—left femur—of a male hunter-gatherer, which was discovered in 2008 protruding from the bank of the Irtysh River by Nikolai Peristov, a Russian sculptor who specialises in carving mammoth ivory. Peristov showed the fossil to a forensic investigator who suggested that it might be of human origin. The fossil was named after the Ust'-Ishim District of Siberia where it had been discovered.
Y-DNA and mtDNA
Ust'-Ishim man belongs to Y-DNA haplogroup K2a*, which is defined by the SNP M2308. The two subclades of K2a known from living men, the rare K2a1-Y28299 and the very common NO-M214, belong to a branch distinct from Ust'-Ishim man's. In the original paper, he was classified only as Haplogroup K2 (the clade ancestral to K2a). It may be inferred that K2a emerged in or near South Asia approximately 47,000 years BP – i.e. K2 is estimated to have originated in South East Asia, about 47,000–55,000 BP, while its secondary descendant NO* is believed to have emerged approximately 38,000 to 47,000 BP. He belonged to mitochondrial DNA haplogroup R*, differing from the root sequence of R by a single mutation.
 |
(Ethnic Thai's are a Mongol people from somewhere in China)
 |
The Mani are the original ethnic group of Thailand. In Thailand they are known as the Sakai, a controversial derogatory term meaning 'slave' or 'barbarism'.They are the only Negrito group in Thailand and speak Maniq (also called Tonga, Kensiu or Mos), a Mon–Khmer language in the Aslian language group. It is thought they once spoke a language similar to the Andamanese language but then adopted the language of the Mon–Khmer people around them. The Maniq are a hunting and gathering society.
The Mani are the most marginalized society on the peninsula today. Pushed out of their preferred lowland forest habitat by logging and agricultural expansion, they now seek refuge – like the last remaining wildlife – in small pockets of protected montane forest. In Malaysia, where they live within Taman Negara National Park, they are known as Orang Asli (Original People), recognizing that a mixture of breeding resulted in the proto Malays, and eventually the Malay peoples of today.
Prior to the southwards migration of the Thai peoples from Guangxi China in the 10th century, mainland Southeast Asia had been home to various indigenous communities for thousands of years. Prior to the arrival of the Thai people and culture into what is now Thailand, the region hosted a number of indigenous Austroasiatic-speaking and Malayo-Sumbawan-speaking civilizations. Thailand was heavily influenced by the culture and religions of India, starting with the Kingdom of Funan around the first century until the Khmer Empire.
 |
(Remembering that full-blood Hawaiians are Black Pacificans, regardless of the fake made-up "Nesian" tags of the Albinos.
According to researchers, Y-DNA (direct paternal lines) in Polynesia has more haplogroup variations than mtDNA (direct maternal lines); however, about 75% reporting their paternal line as Polynesian are in one of the below three Y-DNA haplogroups:
1) Haplogroup C2 [M38]
This is the haplogroup of about 34% who report their paternal line as Polynesian.
C2 is found in Polynesia, Melanesia, New Guinea, and Indonesia.
2) Haplogroup O [M122]
This is the haplogroup of about 24% who report their paternal line as Polynesian.
O is typical of populations of East Asia, Southeast Asia, and culturally Austronesian regions of Oceania [includes Polynesia], with a moderate distribution in Central Asia [ISOGG tree - O3 M122] .
3) Haplogroup K [M9]
This is the haplogroup of about 18% who report their paternal line as Polynesian.
K is an old lineage presently found only at low frequencies in Africa, Asia, and in the South Pacific. One descendent line of this lineage is restricted to aboriginal Australians, while another is found at low frequency in southern Europe, Northern Africa, and the Middle East.
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
Haplogroups L & T (K1)
Haplogroup L (M20) is found in South Asia, Central Asia, South-West Asia, and the Mediterranean.
Haplogroup T (M184, M70, M193, M272) is found at high levels in the Horn of Africa (mainly Cushitic-speaking peoples), parts of South Asia, the Middle East, and the Mediterranean. T-M184 is also found in significant minorities of Sciaccensi, Stilfser, Egyptians, Omanis, Sephardi Jews, Ibizans (Eivissencs), and Toubou. It is also found at low frequencies in other parts of the Mediterranean and South Asia. In India, L-M20 has a higher frequency among Dravidian castes.
Haplogroup K2 (K-M526)
The only living males reported to carry the basal paragroup K2* are indigenous Australians. Major studies published in 2014 and 2015 suggest that up to 27% of Aboriginal Australian males carry K2*, while others carry a subclade of K2.
ISOGG
Geographic distribution so far of ancestry of modern-day haplogroup L subgroup men (not complete): L1a1 M2481 Pakistan, India, Sri Lanka, Bangladesh, Iraq, Bahrain, Kuwait, Saudi Arabia, Qatar,United Arab Em. L1a2 M357 India, Sri Lanka, Bangladesh, Iraq, Kuwait, Saudi Arabia, Qatar, Armenia, Russia, Italy L1b FGC36877/M317 Israel (Druze), Lebanon, Kuwait, Kazakhstan (Tatars), Armenia, Azerbaijan, Iran, Turkey, Belarus, Greece, Bulgaria, Germany, Austria, Switzerland, Portugal, Ukraine, Russia, Mongolia L2 L595 Germany, Sardinian Italy
 |
 |
 |
||
Dravidian |
Uyghur - Turkish-speaking Central Asians in Xinjiang China | Australian |
 |
ISOGG
Y-DNA haplogroup M reaches its known peak in Papua New Guinea, totaling one-third to two-thirds of population.
The haplogroup M Aborigines in Australia seem comparatively to be the most recent arrivals among Aborigines (Bergstrom).
Geographic distribution so far of ancestry of modern-day haplogroup M subgroup men (not complete):
M1b,
M16 Papua New Guinea
M83, Papua New Guinea
M1c,
B271 Papua New Guinea
B269 Papua New Guinea
Z42297 N. Australia (Aborigines)
M2,
M177 Papua New Guinea
M3,
P117 Melanesia
 |
Australians |
 |
Papuan |
 |
Papuan |
 |
Haplogroup N
Haplogroup N (M231) is found through northern Eurasia, especially among speakers of the Uralic languages. Haplogroup N possibly originated in eastern Asia and spread both northward and westward into Siberia, being the most common group found in some Uralic-speaking peoples.
ISOGG
Y-DNA haplogroup N was the predominant paternal haplogroup in northeastern China about 6,500 years before present. It spread widely, but today is most concentrated from the easterrn Baltic across Russia to the Pacific Ocean boundary of Siberia. There is a lesser presence in the southeastern quadrant of Asia, as well as along the East Asia coast. It has an unusual frequency among the more traditional ethnic groups of Russia. The earliest presence recorded in Europe was in Iron Age Hungary. Illumae et al indicate "it remains unclear as to what demographic processes underlie the present-day distribution of hg N.
Geographic distribution so far of ancestry of modern-day haplogroup N subgroup men (not complete):
" N1a1
CTS10760 Finland, Sweden, Sweden (Saami), Norway, Norway (Saami), Estonia, Latvia, Lithuania, Belarus, Ukraine, Poland,
Germany, Czech Republic, Hungary, Romania, Netherlands, England, Scotland
Russia, Russia (Evens, Tatars, Maris, Komis, Mordvins, Chuvashes, Bashkirs, Karanogays)
Russian Siberia (Nenets, Selkups, Tatars, Dolgans)
Z1936 Russia (Bashkirs, Karelians, Vepsas, Cossacks, Yakuts, Bashkirs, Tatars, Karanogays,
Komis, Mordvins, Chuvashes), Russian Siberia (Khanty, Mansi, Nenets, Nganasans, Tatars, Dolgans)
Norway, Sweden, Finland, Estonia, Latvia, Poland, Ukraine, Hungary, Greece, Kazakhstan
B197 Turkey, Ukraine, Mongolia, Kazakhstan, China, China (Tibet), Russia (Tatars, Evenks, Karanogays),
Russian Siberia, Russian Siberia (Buryats, Chuckchis, Koryaks, Eskimos, Tuvans, Altaians, Tatars)
Afghanistan, Uzbekistan, Uzbekistan (Karakalpaks)
M2019 Lebanon, Ukraine, Russian Siberia (Evens, Yakuts, Evenks, Khantys, Tatars, Dolgans, Tuvans, Shorians),
Russia (Yakuts, Karanogays), China, Mongolia, Kazakhstan, Uzbekistan, Uzbekistan (Karakalpaks), Bhutan,
Estonia, Croatia, Turkey
B211 Russia, Russia (Maris, Udmurds, Karelians, Maris, Komis, Mordvins, Chuvashes, Bashkirs, Karanogays), Belarus,
Russian Siberia (Khantys, Altaians, Khakassis)
B187 Russian Siberia (Shorians, Khakas, Khakassis), Russia (Tatars)
N1a2
F1008 Finland, Poland, Belarus, Ukraine, Russia (Vepsas, Maris, Udmurds, Karelians, Komis, Chuvashes, Tatars),
Turkey, Russian Siberia (Evens, Nenets, Evenks, Yakuts, Tuvinians, Khants, Nganasans, Tatars, Dolgans
Eskimos, Tuvans, Khakassis), Armenia, Azerbaijan, Kazakhstan, Afghanistan, Mongolia, China, Japan, Vietnam
N1b
F2930 China, China (Tibet), Japan, Vietnam, India (Telugus), Belarus
N2
P189.2 Serbia, Croatia, Bosnia and Herzegovina, Slovakia"
 |
 |
 |
 |
 |
 |
Haplogroup O
Haplogroup O (M175) is found with its highest frequency in East Asia and Southeast Asia, with lower frequencies in the South Pacific, Central Asia, South Asia, and islands in the Indian Ocean (e.g. Madagascar, the Comoros). Haplogroup O1 (F265/M1354, CTS2866, F75/M1297, F429/M1415, F465/M1422): Haplogroup O1a (M119, CTS31, F589/Page20, L246, L466) Found in eastern,central and southern Mainland China, Taiwan, and Southeast Asia, especially among Austronesian and Tai–Kadai peoples: Haplogroup O1b (P31, M268): Haplogroup O1b1 (M95) Found in Japan, southern China, Taiwan, Southeast Asia, and the Indian subcontinent, especially among Austroasiatic- and Tai–Kadai-speaking peoples, Malays, and Indonesians: Haplogroup O1b2 (SRY465, M176) Found in Japan, Korea, Manchuria, and Southeast Asia: Haplogroup O2 (M122) Found throughout East Asia, Southeast Asia, and Austronesia including Polynesia.
ISOGG
Y-DNA haplogroup O probably originated in East Asia and later migrated into the South Pacific.The lineage expanded into Taiwan (high frequency in the aboriginal Taiwanese), Indonesia, Melanesia, Micronesia, and Polynesia.
O-M122 is the predominant sub-group in China. The O-F265 and O-PK4 lineages are found in Southeast Asian populations of Thailand, Malaysia, Vietnam, Indonesia and southern China. The O-P49 sub-group is of high frequency in the Japanese and Korean populations.
 |
 |
The Mon are the earliest known inhabitants of lower Burma. They founded an empire, and introduced both writing and Buddhism into Burma. In the year 573, two Mon brothers, Prince Samala and Prince Wimala, founded the Mon kingdom Hongsavatoi at the present site of modern Pegu. This kingdom flourished in peace and prosperity for several centuries until it was occupied by the Burman dynasty. In 1757, the Burma ruler U Aungzeya invaded and devastated the Mon kingdom, killing tens of thousands of Mon, including learned Mon priests, pregnant women, and children. Over 3,000 priests were massacred by the victorious Burmans in the capital city alone. Thousands more priests were killed in the countryside. The surviving priests fled to Thailand, and Burman priests took over the monasteries. Most of the Mon literature, written on palm leaves, was destroyed by the Burmans. Use of the Mon language was forbidden, and Burman became the medium of instruction. Mons fled further south into Burma's Tenasserim Division and east into Thailand.
The Khmers, an Austroasiatic people, are one of the oldest ethnic groups in the area, having filtered into Southeast Asia from southern China, possibly Yunnan, around the same time as the Mon, who settled further to the west and to whom the Khmer are ancestrally related. Most archaeologists and linguists, and other specialists like Sinologists and crop experts, believe that they arrived no later than 2000 BCE (over four thousand years ago) bringing with them the practice of agriculture and in particular the cultivation of rice. This region is also one of the first places in the world to use bronze. They were the builders of the later Khmer Empire, which dominated Southeast Asia for six centuries beginning in 802, and now form the mainstream of political, cultural, and economic Cambodia.
 |
 |
The Han Chinese are an East Asian ethnic group native to China. They constitute the world's largest ethnic group, making up about 18% of the global population and consisting of various subgroups speaking distinctive varieties of Chinese languages. The estimated 1.3 billion Han Chinese people are mostly concentrated in mainland China, where they make up about 92 percent of the total population. In Taiwan, they make up about 97% of the population. People of Han Chinese descent also make up around 75% of the total population of Singapore.
The Han Chinese show a close genetic relationship with other modern East Asians such as the Koreans and Yayoi of Japan (now called Yamato - Modern Japanese). Y-chromosome haplogroup O2-M122 is a common DNA marker in Han Chinese, it is found in at least 36.7% to over 80% of Han Chinese males in certain regions. Other Y-DNA haplogroups that have been found with notable frequency in samples of Han Chinese include O-P203 (9.1%), C-M217 (6.0%), N-M231 (3.6%), O-M268 (4.7%), and Q-M242 (1.2%).
The main Mongol Y-haplogroups of Japan are O1 and O2. A genome research (Takashi et al. 2019) confirmed that modern Japanese (Yamato) descend mostly from the Yayoi people who came from China. Mitochondrial DNA analysis of Jōmon and modern Japanese samples show that there is a discontinuity between the mtDNAs of people from the Jōmon period and people from the Kofun and Heian periods. Recent studies have revealed that Jomon people are considerably genetically different from any other population, including modern-day Japanese. Takahashi et al. 2019, (Adachi et al., 2011; Adachi and Nara, 2018). The population found to be closest to the Jōmon was the Ainu.
Chinese Woman |
Japanese Woman |
Korean Woman |
ISOGG
Y-DNA haplogroup P, an offshoot of Haplogroup K2b, originated in Central or Eastern Asia some 35,000 years ago. Haplogroup P is best represented by its two immediate subclades, haplogroups Q and R, which expanded to become the dominant haplogroups in, respectively, the Americas and Europe. P1-M45 has been found in n. Philippines, India, China (Maks, Ai Cham, Biao, Then, Uygurs, Tibetans, Hans), Taiwan (Pyuma), Indonesia (Batak, Malay, Minangkabau, Kaili, Alor), Iran (Bakhtiari, Arabs) Pakistan (Burushos), Melanesia, Jordan, Croatia, Romania (Szeklers), Scandinavia, (It is not clear that all these men were verified negative for the haplogroup Q subgroup). P2-B253 was identified in the Philippines (Agtas)
Haplogroup P (K2b2)
Basal P* is found at its highest rate among members of the Aeta (or Agta), a people indigenous to Luzon, in The Philippines. Luzon is also the only location where P*, P1* and rare P2 are now found together, along with significant levels of K2b1. Basal P* is also found in one historical 19th-century Andaman islander. Even though P1* is now more common among individuals in Eastern Siberia and Central Asia, the above distributions tend to suggest that P* (P295) emerged in South East Asia. P1 is also the parent node of two primary clades: Haplogroup Q (Q-M242) and; Haplogroup R (R-M207). These share the common marker M45 in addition to at least 18 other SNPs.
 |
Aeta people Philippines |
 |
Eastern Siberian |
 |
Eastern Siberian - Ket people |
 |
ISOGG
Y-DNA haplogroup Q arose in Central Asia and migrated through the Altai/Baikal region of northern Eurasia into the Americas. Today it is found in North Eurasia, with some exemplars in European populations. The Q-M3 sub-group is almost exclusively associated with Native American populations.
Geographic distribution so far of ancestry of modern-day haplogroup Q subgroup men (not complete): Vietnam, Russian Siberia (Koryats), Indonesia (Muruts), Bhutan, Russia (F746 ancient DNA, c16700 yrs ago-Afontova Gora 2 & 3) ago-Afontova Gora 2 & 3), Uzbekistan (Turkmens), Pakistan (Punjabis), Russia, Poland, Hungary, UK, Bahrain, Turkey, England, Sweden, France
CTS11969/M930 group
[M3 subgrp of CTS11969 earlier than these subgrps below, found
as M3+ ancient DNA, Kennewick man, Washington state, USA, lived abt 9000 yrs ago],
M19 S. America (Ticunas, Wayuus, Tobas),
SA01 Peru, Bolivia,
CTS11357 Colombia, Nicaragua, Guatemala, El Salvador, Mexico, Brazil (Karitianas),
Z35840 Argentina (Cachis), Peru, PF1337.2 Peru,
Z5906 Peru, Argentina (Collas, Cachis),
Z5908 Peru, Argentina (Collas, Cachis),
Y789 Peru, Argentina (Collas),
Z19432 Peru, Argentina (Collas),
SK281 Peru,
CTS2731 Mexico,
Y27992 Mexico, Argentina,
CTS133 Mexico,
B43 Argentina (Wichis),
Z35706 Peru,
CTS44 Colombia,
Z35737 Peru,
Z35742 Peru,
Z35747 Peru,
BZ3401 Brazil,
A1211 s.e. USA, n.e. USA (Wappingers, Mohicans),
FGC8469 Russian Siberia (Eskimos), n.e. USA (Wampanoags). s.e. USA, Mexico
M971/Z780 group;
L569 USA,
Z782 Mexico,
L456 central USA (Sioux),
L191 Mexico,
BY13107 s.w. USA,
E324/L804 Sweden, Norway, Iceland, Ireland, Denmark, Germany, England, France,
L330 Kazakhstan, Uzbekistan, France, Russian Siberia (Selkups, Kets),
BZ577 Poland, Russia,
B28 Nepal, UK, Sweden, Norway, Finland, Bosnia & Herzegovina, Croatia, Sardinian Italy, Germany, Netherlands, Portugal, Belarus, United Arab Emirates, Yemen (Jews), Pakistan (Pathans), Sri Lanka (Tamils), India (Syrian Christians)
Q2
FGC1917 Poland, Italy, Ukraine, n. e. Europe (including Ashkenazi Jews), China (Karaites),
YP745 Morocco, Jordan, Iraq, Turkey (Armenians, Assyrians), Switzerland, Russia, Russia (Avars), China (Ughurs), India,
BZ310 Iran, Azerbaijan,
BZ3896 Italy, Armenia,
FGC4607 Portugal, Kazakhstan, India, Pakistan (Punjabis), Iran (Arabs), Georgian Abkhazia,
L68-Y1143 Pakistan (Punjabis), Russia (Tatars), Bangladesh, Poland
 |
 |
|||
La Brea Woman |
Pelican Rapids-Minnesota Woman |
Spirit Cave Mummy |
||
| Los Angeles' famed La Brea Tar Pits: La Brea Woman is the name for a human whose remains were found in the La Brea Tar Pits in Los Angeles, California. The remains, first discovered in the pits in 1914, were the partial skeleton of a woman. At around 18-25 years of age at death, she has been dated at 10,220 years old. | Pelican Rapids-Minnesota Woman, is the skeletal remains of a woman thought to be 8,000 years old. The bones were found near Pelican Rapids, Minnesota on June 16, 1931. The University of Minnesota identified them as the bones of a woman who was 15 or 16 years old. | The skeleton of 10,600-year-old man shrouded in a rabbit-skin blanket and reed mats is the oldest known mummy ever found in the United States. It was discovered in 1940 in Spirit Cave, 13 miles (21 km) east of Fallon, Nevada. The mummy is older than any previously known North American mummy. |
 |
The problem is that Anzick child and Kennewick man are only two of many that have been genetically typed.
The reason they have not been genetically typed is because of resistance from "Modern" so-called Indians.
Who as it turns out are Albinos or the Mulattoes of Albinos.
The Albinos in the U.S. government gave "Other" Albinos, and their Mulattoes "CONTROL" over all Indian affairs.
Loring Brace (University of Michigan) and Richard Jantz (University of Tennessee, Knoxville) are attempting to incorporate cranio-facial measurements from Pacific Northwest crania into their respective worldwide comparative databases. Jantz and Owsley (1999a) are performing multivariate analyses to explore differences between ancient crania and modern populations. They have recently argued that Buhl skeletal remains show differences between the ancient and modern populations, and that Buhl's morphometric traits are not similar to modern Native American groups; in fact they are closer to groups from the Pacific. They suggest that a source of the early migrants to America might be found in Asian Circumpacific populations. These populations are quite naturally variable, but their craniofacial morphology consists of cranial vaults that are large, long and narrow, forward projection of the face, and low faces. Polynesians and some ancestors to early California Indian populations probably came out of these populations. More recently Jantz and Owsley (2000) analyzed a sample of 11 crania (Spirit Cave, Wizards Beach, Browns Valley, Pelican Rapids, Prospect, Wet Gravel male, Wet Gravel female, Medicine Crow, Turin, Lime Creek, and Swanson Lake). The sample includes the pre-Mazama Prospect burial, from Oregon. As early as 1991 Brace and his collaborators (Brace et al. 1990) began to suggest that their multivariate analysis of the world-wide Michigan database showed that west coast Amerindian samples most closely aligned with the Jomon-Pacific samples.
Neves and Blum (2000) are testing the recent claim that craniofacial observations of the Buhl Paleoindian remains are similar to other North American and East Asian populations. The measurements of the Buhl skull were compared to twenty-six modern populations (Howells), and to a Paleoindian skull from Lapa Vermelha, Brazil, which shows morphological similarities with Africans and Australians. Multivariate analysis shows that there is a great difference between the Paleoindian skulls, and when compared to the modern populations the skulls belong to different clusters. They suggest that at least two populations peopled the Americas; one with characteristic "Mongoloid morphology," and another with a generalized morphology. |
 |
 |
 |
Haplogroup Q is the major branch on the Y-chromosome tree (89%) in modern population set.
Maya Populations: Mitochondrial DNA Haplogroups A, B, C, and D
DNA studies and the origin of the Olmecs (2018), by Enrique Villamar Becerril
The study of ADNMT carried out on Olmec individuals, one from San Lorenzo and the other from Loma del Zapote, resulted, in both cases, in the unequivocal presence of the distinctive mutations of the “A” maternal lineage.
 |
 |
Late Ceramic Guadeloupeans (LCG) DNA was obtained from 13 human remains, out of the 38 tested, distributed over the entirety of the Guadeloupe archipelago, and dating from between AD 1200 and 1600.
The distribution of the mitochondrial haplogroups in the pre-Columbian LCG group was as follows: 38.5% A2, 38.5% C1 and 23% D1. The B2 mitochondrial haplogroup was not detected in the ancient group. In addition, eight Y-chromosomal SNPs were also amplified to characterize the major Amerindian paternal lineages: C-M216, Q-M242, Q-M346, Q-M3, Q-M19, Q-M194, Q-M199 and Q-SA01. The mitochondrial gene pool of the group was characterized by the presence of three Amerindian mitochondrial haplogroups (A2, C1 and D1).
As illustrated in figure 2b, the same pattern of genetic affinities was observed when we considered all of the ancient Caribbean samples. The ancient Caribbean groups could not be significantly differentiated from native Colombians inhabiting the Amazon area and they were only slightly differentiated from the Amazonian and Colombian populations. The PCA analysis, based on the haplogroup frequencies, confirmed the affinity of the LCG group with some South American groups (such as the Apalai, Guahibo or Arawak), especially owing to the high percentages of C1 haplogroup Native American Haplogroups of the Caribbean, Mesoamerica, and South America.
Only weak genetic affinities were observed between the ancient Caribbean groups, including our LCG sample, and the extant populations of Mesoamerica. This observation underscores the low amount of gene flow between both regions during the ceramic periods. However, it should be noted that two rare mitochondrial haplotypes (Ht-04 and Ht-06) were shared between the LCG group and a few native individuals from Mexico and Honduras (Garifunas). The Garifunas, or ‘Black Caribs’.
From the study: Reconstructing the Population Genetic History of the Caribbean (2013)
Population structure of the Caribbean
To characterize population structure across the Antilles and neighboring mainland populations, we combined our genotype data for the six Latino populations with continental population samples from western Africa, Europe, and the Americas, as well as additional admixed Latino populations. To maximize SNP density, we initially restricted our reference panels to representative subsets of populations with available Affymetrix SNP array data. Using a common set of 390 K SNPs, we applied both principal component analysis (PCA) and an unsupervised clustering algorithm, ADMIXTURE, to explore patterns of population structure. Figure 1B shows the distribution in PCA space of each individual, recapitulating clustering patterns previously observed in Hispanic/Latino populations: Mexicans cluster largely between European and Native American components, Colombians and Puerto Ricans show three-way admixture, and Dominicans principally cluster between the African and European components. Ours is the first study to characterize genomic patterns of variation from (1) Hondurans, which we show have a higher proportion of African ancestry than Mexicans, (2) Cubans, which show extreme variation in ancestry proportions ranging from 2% to 78% West African ancestry, and (3) Haitians, which showed the largest average proportion of West African ancestry (84%).
 |
Haplogroup R, or R-M207 is both numerous and widespread amongst modern populations. Some descendant subclades have been found since pre-history in Europe, Central Asia and South Asia. Others have long been present, at lower levels, in parts of West Asia and Africa. Some authorities have also suggested, more controversially, that R-M207 has long been present among Native Americans in North America – a theory that has not yet been widely accepted. Haplogroup R* Y-DNA (xR1,R2) was found in 24,000-year-old remains from Mal'ta in Siberia near Lake Baikal. In 2013, R-M207 was found in one out of 132 males from the Kyrgyz people of East Kyrgyzstan.
The earliest human occupation in this region probably began sometime around 40,000 years ago. Small groups of big-game hunters likely migrated into this region from lands to the south and southwest, confronting a harsh climate and long, dry winters. By about 20,000 B.C., two principal cultural traditions had developed in Siberia and northeastern Asia: the Mal’ta and the Afontova Gora-Oshurkovo.
The Mal’ta tradition is known from a vast area spanning west of Lake Baikal and the Yenisey River. The site of Mal’ta, for which the culture is named, is composed of a series of subterranean houses made of large animal bones and reindeer antler which had likely been covered with animal skins and sod to protect inhabitants from the severe, prevailing northerly winds. Among the artistic accomplishments evident at Mal’ta are remains of expertly carved bone, ivory, and antler objects. Figurines of birds and human females are the most commonly found items.
The Mal'ta–Buret' culture is an archaeological culture of the Upper Paleolithic (c. 24,000 to 15,000 BP) on the upper Angara River in the area west of Lake Baikal in the Irkutsk Oblast, Siberia, Russian Federation. The type sites are named for the villages of Mal'ta, Usolsky District and Buret', Bokhansky District (both in Irkutsk Oblast).
A boy whose remains were found near Mal'ta is usually known by the abbreviation MA-1 (or MA1). Discovered in the 1920s, the remains have been dated to 24,000 BP. According to research published since 2013, MA-1 belonged to a population related to the genetic ancestors of Siberians, American Indians, and Bronze Age Yamnaya and Botai people of the Eurasian steppe. In particular, modern-day Native Americans, Kets, Mansi, and Selkup have been found to harbor a lot of ancestry related to MA-1.
MA-1 is the only known example of basal Y-DNA R* (R-M207*) – that is, the only member of haplogroup R* that did not belong to haplogroups R1, R2 or secondary subclades of these. The mitochondrial DNA of MA-1 belonged to an unresolved subclade of haplogroup U.
 |
 |
 |
 |
 |
 |
 |
 |
||
R-M173 - also known as R1, has been common throughout Europe and South Asia since pre-history. It has many branches, It is the second most common haplogroup in Indigenous peoples of the Americas following haplogroup Q-M242, especially in the Algonquian peoples of Canada and the United States. The reasons for high levels of R-M173 among Native Americans are a matter of controversy: authorities point to the greater similarity of many R-M173 subclades found in North America to those found in Siberia, suggesting prehistoric immigration from Asia and/or Beringia.
As an abject lesson of how racism isrelated to Power: note the following.
By the time the Albino British had left India, the docile and impressionable Mulatto Hindu's had accepted Albinos as demigods. Even the supposed great humanist Mohandas Karamchand Gandhi wrote this in 1903 when Gandhi was in South Africa. He wrote that white people there should be "the predominating race." He also said black people "are troublesome, very dirty and live like animals."
When Albinos invade and colonize, they produce Mulattoes. Those Mulattos who accept their Albino parent as superior, and identify with them, are given advancement. Then when the Albinos are forced to leave, it is their Albino worshipping Mulattos who have the education and training to take over government. Thus lands with significant Mulatto populations like India, the Middle East, and Latin America, have very anti-Black cultures.Today, in spite of being, by HUGE majority, peopled by BLACK and BROWN skinned peopled: India, is considered the most RACIST and Pro-Albino country in the ENTIRE WORLD! Sick, sick, sick. An entire nation with "Stockholm Syndrome". |
 |
 |
Haplogroup R (M207)
The hypothetical divergence of Haplogroup R and its descendants. Haplogroup R is defined by the SNP M207. The bulk of Haplogroup R is represented in descendant subclade R1 (M173), which likely originated on the Eurasian Steppes. R1 has two descendant subclades: R1a and R1b. R1a is associated with the proto-Indo-Iranian and Balto-Slavic peoples, and is now found primarily in Central Asia, South Asia, and Eastern Europe.
Haplogroup R1b is the dominant haplogroup of Western Europe and also found sparsely distributed among various peoples of Asia and Africa. Its subclade R1b1a2 (M269) is the haplogroup that is most commonly found among modern Western European populations, and has been associated with the Italo-Celtic and Germanic peoples.
Haplogroup R1 (M173) Found throughout western Eurasia: Haplogroup R1a (M420) Found in Central Asia, South Asia, and Central, Northern and Eastern Europe: Haplogroup R1b (M343) Found in Western Europe, West Asia, Central Asia, North Africa, and northern Cameroon,
Haplogroup R2 (M124) Found in South Asia, Caucasus, Central Asia, and Eastern Europe
ISOGG
"Y-DNA haplogroup R-M207 is believed to have arisen approximately 27,000 years ago in Asia. The two currently defined subclades are R1 and R2.
R1-M173 is estimated to have arisen during the height of the Last Glacial Maximum (LGM), about 18,500 years ago, most likely in southwestern Asia. The two most common descendant clades of R1 are R1a and R1b.
R1a-M420 is believed to have arisen on the Eurasian Steppe or the Indus Valley, and today is most frequently observed in eastern Europe and in western and central Asia. R1a-M458 is found at frequencies approaching or exceeding 30% in Eastern Europe.
R1b-M343 is believed to have arisen in southwest Asia and today its sublcades are bound in various distributions across Eurasia and Africa.
Paragroup R1b1* and R1b1b-V88 are found most frequently in SW Asia and Africa. The African examples are almost entirely within R1b1b and are associated with the spread of Chadic languages.
R1b1a1-P297 is found throughout Eurasia. R1b-M73 is observed most frequently in Asia, with low frequency of observation in Europe. R1b-M269 is observed most frequently in Europe, especially western Europe, but with notable frequency in southwest Asia. R1b1a1b-M269 is estimated to have arisen approximately 4,000 to 8,000 years ago in southwest Asia and to have spread into Europe from there. The Atlantic Modal Haplotype, or AMH, is the most common STR haplotype in R1b1a1b1a1a-L11/PF6539/S127 and most European R1b belongs to R1b1a1b1a1a1-M405/S21/U106 or R1b1a1b1a1a2-P312/PF6547/S116. R2-M479 is most often observed in Asia, especially on the Indian sub-continent and in central Asia."
Y-haplogroup R1b
Africa - Cameroon 95%, Berber 28%, Niger 20%, Central Sahel Region 23%, North Africa 6%, Western Europeans 58%, North-west Europeans 56%, Central Europeans 43%, Eastern Europeans 21%, West Asians 6%.
 |
 |
 |
||
Cameroon |
Berber |
Central Sahel |
Western Europe includes: Austria, Belgium, Czech Republic, France, Germany, Ireland, Liechtenstein, Luxembourg, Monaco, Netherlands, Switzerland, United Kingdom.Eastern Europe, includes Greece, Belarus, Bulgaria, Serbia, Romania, Russia, and Ukraine. Geographically, Northwestern Europe usually consists of the United Kingdom, the Republic of Ireland, Belgium, the Netherlands, Luxembourg, Northern France, Northern Germany, Denmark, Norway, Sweden, and Iceland.
 |
 |
 |
||
British |
German |
Russian |
Y-haplogroup R1a
In India/Asia: West Bengal Brahmins (72%), Gujarat Lohanas (60%), Khatris (67%), Manipuris (50%), Punjabis (47%), Pakistan 71%, Terai region of Nepal 69%. Black-Iranians (Persians),18.4%.
 |
 |
 |
||
Brahmin |
Lohanas |
Khatris |
 |
 |
 |
||
Pakistan |
Nepal |
Iran |
In Europe: the R1a1 sub-clade is found at highest levels among peoples of Central and Eastern European descent, with results ranging from 35-65% among Czechs, Hungarians, Poles, Slovaks, western Ukrainians (particularly Rusyns), Belarusians, Moldovans, and Russians. In the Baltics, R1a1a frequencies decrease from Lithuania (45%) to Estonia (around 30%). There is a significant presence in peoples of Scandinavian descent, with highest levels in Norway and Iceland, where between 20 and 30% of men are in R1a1a. In East Germany, where Haplogroup R1a1a reaches a peak frequency in Rostock at a percentage of 31.3%, it averages between 20 and 30%. In Southern Europe R1a1a is not common, but significant levels have been found in pockets, such as in the Pas Valley in Northern Spain, areas of Venice, and Calabria in Italy. The Balkans shows lower frequencies, and significant variation between areas, for example more than 30% in Slovenia, Croatia and Greek Macedonia, but less than 10% in Albania, Kosovo and parts of Greece on south from Olympus gorge. R1a is virtually composed only of the Z284 subclade in Scandinavia.
 |
 |
 |
||
Czech |
Hungarian |
Pole |
 |
ISOGG
Y-DNA haplogroup S is a major haplogroup in the highlands of mainland Papua New Guinea where it is found at frequencies of around 50% in some populations and is also present at lower frequencies in adjacent islands of Indonesia and Melanesia and even in the Philippines.
Geographical Distribution of ancestry of men in Haplogroup S (not complete):
S1a1 Z42413 Indonesia,
S1a1a1 P60 Australia (Aborigines) P60 Australia (Aborigines), S1a1b1d1a M226.1 Papua New Guines, Admiralty Islands M226.1 Papua New Guinea, Admiralty Islands,
S1a2 P79 Melanesia, Papua New Guinea including Admiralty Islands,
S1a3 P315 Australia (Aborigines),
S1a3b P401,
S1b B275 Philippines (Bajos),
S2 B273 Philippines (Aeta),
S3 P336 Indonesia,
S4 BY22870 Philippines
 |
 |
|
Batek Tribe Philippines |
Malaysia |
 |
Papua mountain people |
 |
 |
|
Tamil People |
Tamil People |
As is apparent in this presentation, Asia was the original Melting Pot of Man. A good example of a thoroughly Mixed (Mulatto) population are the Karakalpak nation of Uzbekistan.
The Karakalpaks/Qaraqalpaqs are a Turkic speaking ethnic group native to Karakalpakstan in Northwestern Uzbekistan. During the 18th century, they settled in the lower reaches of the Amu Darya and in the (former) delta of Amu Darya on the southern shore of the Aral Sea. The name "Karakalpak" comes from two words: "qara" meaning black and "qalpaq" meaning hat. The Karakalpaks number nearly 620,000 worldwide, out of which about 500,000 live in the Uzbek Republic of Karakalpakstan.
Recent archaeological evidence indicates that the Karakalpaks may have formed as a confederation of different tribes at some time in the late 15th or the 16th centuries at some location along the Syr Darya or its southern Zhany Darya outlet, in proximity to the Kazakhs of the Lesser Horde. This would explain why their language, customs and material culture are so similar to that of the Kazakhs.
Karakalpaks are primarily followers of the Hanafi School of Sunni Islam. It is probable they adopted Islam between the 10th and 13th centuries, a period when they first appeared as a distinct ethnic group. Dervish orders such as the Naqshbandi, Kubrawiya, Yasawi and Qalandari are fairly common in this region. The religious order that established the strongest relation with the people of the region is the Kubrawiya, which has Shi'i adherents.
Y-dna Haplogroups present in Karakalpak nation in Uzbekistan
J-M172
O-M175
I-P37
Q-M242, Q-L53
G-Z6638, G-M201
N-M232, N-M232
C-M216, C-M217, C-Y20087, C-F4002, C-F5481
R-M343, R-M512, R-M173, R-M124, R-BY42301
Karakalpak nation People |
 |
In an effort to bring sanity tothe delusional minds of Albinos
From early on, we Afrocentrics realized that the minds of Western Albinos were shaped by western media: i.e. Movies, Television, Books, Magazines, etc. (note our main page). Black people think it so ridiculous to see an Albino Man named Tarzan, swinging through the trees in Africa. Now Albino media has a program titled "Naked and Afraid" were we see Albinos totally Naked, walking around in Africa, Central America, the Southern United States, and other such areas: and Albinos think it perfectly plausible! And despite seeing pictures of Black skinned ancient Egyptians working their fields "SHIRTLESS", which should have given them a clue: because of other clear evidence in their own lives: i.e. how easily they Sunburn, get Skin Cancer, etc. Albinos think that perfectly plausible too. All because Albino Movies and Television said that Albinos were Ancient Egyptians, Persians, Mesopotamian's; and also that they could walk around naked in high UV environments. And so armed with what Movies and T.V. said, Western Albinos blindly believed that indeed, people like them could have been Egyptians, Persians, Mesopotamian's, Tarzan: and they can walk around in those high UV areas Naked, without lathering on SPF 100 Sunscreen every 15 min. and even with that: Medical Science tells them to seek shelter in UV index 10+ areas. They also tell them "Do Not Burn" Five or more sunburns doubles your risk of developing skin cancer! As a point of fact, the average July temperature in Jackson Mississippi is about 91 degrees, and the UV is 10 (the max is 11). But now we have happened across a rather obscure document; specifically the declaration of Secession of the State of Mississippi. These Mississippians like the other Slavers were degenerate barely Human Creatures, who were not even embarrassed by the fact that they were so fragile in body, that they were not able to go out into the Sunlight and do basic Farming, so that they might FEED themselves. So they honestly said WHY they NEEDED Slaves. And as is typical with people with a disability that cannot be repaired, they even try to make it sound like it's a benefit and that they are proud of it. Leaving their degeneracy and delusion to them: the point is that the document was written almost a hundred years before Movies and Television existed, therefore it was written before the FANTASY'S created by Movies and T.V. existed. So for those Albinos wanting to touch REALITY again, we offer the Secession Declaration of Mississippi.
|
Repeating - the Albinos lie, lie, lie, when it comes to the ancient Egyptians, Greeks, and Romans. For those who know the facts it's rather pathetic, but to those ignorant of the facts, they can be persuasive. Therefore....
By Ricaut FX1, Waelkens M. (2008)
Harran is mentioned in the Hebrew Bible, it first appears in the Book of Genesis as the home of Terah and his descendants, and as Abraham's temporary home. |
Quote: We know from archeological data that in the upper Paleolithic period Anatolia was settled by populations with Aurignacian culture (Kuhn 2002). Recent genetic studies (Cinnioglu et al. 2004; Olivieri et al. 2006) based on the analysis of mtDNA (haplogroup M1 and U6) and the Y chromosome (R1b3-M269 lineage) suggest, in agreement with paleoenvironmental evidence (van Andel and Tzedakis 1996), that around 40,000–45,000 years ago, populations with Aurignacian culture may have spread by migration from the Levant and southwest Asia to Anatolia and further into Europe (Bar-Yosef 2002). With the exception of these scarce molecular data, almost nothing is known about the biological features of these early Paleolithic Anatolian foragers. Nevertheless, considering the important demographic processes and biological changes undergone by human populations as a result of later and major events (e.g., the Neolithic transition), we believe that the causes of the observed affinity patterns have to be determined from these later periods.
From the Mesolithic to the early Neolithic period different lines of evidence support an out-of-Africa Mesolithic migration to the Levant by northeastern African groups that had biological affinities with sub-Saharan populations.
{The Levant = Cyprus, Israel, Iraq, Jordan, Lebanon, Palestine, Syria, southern Turkey}.
From a genetic point of view, several recent genetic studies have shown that sub Saharan genetic lineages (affiliated with the Y-chromosome PN2 clade; Underhill et al. 2001) have spread through Egypt into the Near East, the Mediterranean area, and, for some lineages, as far north as Turkey (E3b-M35 Y lineage; Cinnioglu et al. 2004; Luis et al. 2004), probably during several dispersal episodes since the Mesolithic (Cinnioglu et al. 2004; King et al. 2008; Lucotte and Mercier 2003; Luis et al. 2004; Quintana-Murci et al. 1999; Semino et al. 2004; Underhill et al. 2001).
This finding is in agreement with morphological data that suggest that populations with sub-Saharan morphological elements were present in northeastern Africa, from the Paleolithic to at least the early Holocene, and diffused northward to the Levant and Anatolia beginning in the Mesolithic.
Indeed, the rare and incomplete Paleolithic to early Neolithic skeletal specimens found in Egypt—such as the 33,000-year-old Nazlet Khater specimen (Pinhasi and Semal 2000), the Wadi Kubbaniya skeleton from the late Paleolithic site in the upper Nile valley (Wendorf et al. 1986), the Qarunian (Faiyum) early Neolithic crania (Henneberg et al. 1989; Midant-Reynes 2000), and the Nabta specimen from the Neolithic Nabta Playa site in the western desert of Egypt (Henneberg et al. 1980)—show, with regard to the great African biological diversity, similarities with some of the sub-Saharan middle Paleolithic and modern sub-Saharan specimens.
This affinity pattern between ancient Egyptians and sub-Saharans has also been noticed by several other investigators (Angel 1972; Berry and Berry 1967, 1972; Keita 1995) and has been recently reinforced by the study of Brace et al. (2005), which clearly shows that the cranial morphology of prehistoric and recent northeast African populations is linked to sub-Saharan populations (Niger-Congo populations). These results support the hypothesis that some of the Paleolithic–early Holocene populations from northeast Africa were probably descendents of sub-Saharan ancestral populations.
A late Pleistocene–early Holocene northward migration (from Africa to the Levant and to Anatolia) of these populations has been hypothesized from skeletal data (Angel 1972, 1973; Brace et al. 2005) and from archeological data, as indicated by the probable Nile valley origin of the “Mesolithic” (epi-Paleolithic) Mushabi culture found in the Levant (Bar Yosef 1987). This migration finds some support in the presence in Mediterranean populations (Sicily, Greece, southern Turkey, etc.; Patrinos et al. 2001; Schiliro et al. 1990) of the Benin sickle cell haplotype. This haplotype originated in West Africa and is probably associated with the spread of malaria to southern Europe through an eastern Mediterranean route (Salares et al. 2004) following the expansion of both human and mosquito populations brought about by the advent of the Neolithic transition (Hume et al. 2003; Joy et al. 2003; Rich et al. 1998).
This northward migration of northeastern African populations carrying sub-Saharan biological elements is concordant with the morphological homogeneity of the Natufian populations (Bocquentin 2003), which present morphological affinity with sub-Saharan populations (Angel 1972; Brace et al. 2005). In addition, the Neolithic revolution was assumed to arise in the late Pleistocene Natufians and subsequently spread into Anatolia and Europe (Bar-Yosef 2002), and the first Anatolian farmers, Neolithic to Bronze Age Mediterraneans and to some degree other Neolithic–Bronze Age Europeans, show morphological affinities with the Natufians (and indirectly with sub-Saharan populations; Angel 1972; Brace et al. 2005), in concordance with a process of demic diffusion accompanying the extension of the Neolithic revolution (Cavalli-Sforza et al. 1994).
Following the numerous interactions among eastern Mediterranean and Levantine populations and regions, caused by the introduction of agriculture from the Levant into Anatolia and southeastern Europe (Bar-Yosef 2002; Keita and Boyce 2005; King et al. 2008), there was, beginning in the Bronze Age, a period of increasing interactions in the eastern Mediterranean, mainly during the Greek, Roman, and Islamic periods. These interactions resulted in the development of trading networks, military campaigns, and settler colonization (Cruciani et al. 2007; Edwards et al. 2000; Keita and Boyce 2005; King et al. 2008; Lucotte and Mercier 2003; Sahoglu 2005; Waelkens et al. 2006).
Major changes took place during this period, which may have accentuated or diluted the sub-Saharan components of the earlier Anatolian populations. The second option seems more likely, because even though the population from the Sagalassos territorywas interacting with northeastern African and Levantine populations [trade relationships with Egypt (Arndt et al. 2003), involvement of thousands of mercenaries from Pisidia (Sagalassos region) in the war around 300 b.c. between the Ptolemaic kingdom (centered on Egypt) and the Seleucid kingdom (Syria/Mesopotamia/Anatolia), etc.], the major cultural and population interactions involving the Anatolian populations since the Bronze Age occurred with the Mediterranean populations from southeastern Europe, as suggested from historical (cf. historical context) and genetic data (Berkman et al. 2008; Cinnioglu et al. 2004; Di Benedetto et al. 2001; Tambets et al. 2000).
Consequently, one may hypothesize as the most parsimonious explanation that sub-Saharan biological elements were introduced into the Anatolian populations after the Neolithic spread and have been preserved since this time, at least until the 11th–13th century a.d, in the population living in the Sagalassos territory of southwestern Anatolia.
This scenario implies that the affinity between Sagalassos and the two sub-Saharan populations (Gabon and Somalia) is more likely due to the sharing of a common ancestor and that the major changes and increasing interactions in the eastern Mediterranean beginning in the Bronze Age did not erase some of the sub-Saharan elements carried by Anatolian populations, as shown by genetic data (e.g., Cinnioglu et al. 2004; Luis et al. 2004) and the morphological features of our southwestern Anatolian sample.
Abstract
We study the major levels of Y-chromosome haplogroup variation in 15 Sudanese populations by typing major Y-haplogroups in 445 unrelated males representing the three linguistic families in Sudan. Our analysis shows Sudanese populations fall into haplogroups A, B, E, F, I, J, K, and R in frequencies of 16.9, 7.9, 34.4, 3.1, 1.3, 22.5, 0.9, and 13% respectively. Haplogroups A, B, and E occur mainly in Nilo-Saharan speaking groups including Nilotics, Fur, Borgu, and Masalit; whereas haplogroups F, I, J, K, and R are more frequent among Afro-Asiatic speaking groups including Arabs, Beja, Copts, and Hausa, and Niger-Congo speakers from the Fulani ethnic group. Mantel tests reveal a strong correlation between genetic and linguistic structures (r = 0.31, P = 0.007), and a similar correlation between genetic and geographic distances (r = 0.29, P = 0.025) that appears after removing nomadic pastoralists of no known geographic locality from the analysis. The bulk of genetic diversity appears to be a consequence of recent migrations and demographic events mainly from Asia and Europe, evident in a higher migration rate for speakers of Afro-Asiatic as compared with the Nilo-Saharan family of languages, and a generally higher effective population size for the former. The data provide insights not only into the history of the Nile Valley, but also in part to the history of Africa and the area of the Sahel. Am J Phys Anthropol, 2008. © 2008 Wiley-Liss, Inc.
Results of haplogroup diversity
Haplogroup frequencies in 15 Sudanese populationsare given in Figure 2 following YCC nomenclature(2002). Haplogroups A-M13 and B-M60 are present at high frequencies in Nilo-Saharan groups except Nubians, with low frequencies in Afro-Asiatic groups although no-table frequencies of B-M60 were found in Hausa (15.6%) and Copts (15.2%). Haplogroup E (four different haplo-types) accounts for the majority (34.4%) of the chromosome and is widespread in the Sudan. E-M78 represents 74.5% of haplogroup E, the highest frequencies observed in Masalit and Fur populations. E-M33 (5.2%) is largely confined to Fulani and Hausa, whereas E-M2 is restricted to Hausa. E-M215 was found to occur more in Nilo-Saharan rather than Afro-Asiatic speaking groups. In contrast, haplogroups F-M89, I-M170, J-12f2, and J-M172 were found to be more frequent in the Afro-Asiatic speaking groups.
J-12f2 and J-M172 represents 94% and 6%, respectively, of haplogroup J with high frequencies among Nubians, Copts, and Arabs. Haplogroup K-M9 is restricted to Hausa and Gaalien with low frequencies and is absent in Nilo-Saharan and Niger-Congo. Haplogroup R-M173 appears to be the most frequent haplogroup in Fulani, and haplogroup R-P25 has the highest frequency in Hausa and Copts and is present at lower frequencies in north, east, and western Sudan. Haplogroups A-M51, A-M23, D-M174, H-M52, L-M11, O-M175, and P-M74 were completely absent from the populations analyzed .Fig. 1. Map of the Sudan. Showing the approximate locations of 15 populations typed for Y-chromosome markers in this study. The nomadic populations, Meseria, and Fulani were not shown in the map due to their wide distribution in west, east, and south-west of the country. Circles indicate the geographic regions. Arrows indicate directions of assigned location to population in Mantel Test, based on knowledge of history of migration within the past hundred years.317 Y-CHROMOSOME VARIATION AMONG SUDANESE American Journal of Physical Anthropology
Misseriya Arabs, are a branch of the Baggara ethnic grouping of Arab tribes. Numbering over one million, the Baggara are the second largest ethnic group in Western Sudan. They are primarily nomadic cattle herders. The use of the term Baggara carries negative connotations as slave raiders, so they prefer to be called Messiria instead.
Based on cultural traits, many scholars believe Nubia is derived from the Ancient Egyptian: nbw "gold".
The Roman Empire used the term "Nubia" to describe the area of Upper Egypt and Northern Sudan.
Pure Nubians are an ethno-linguistic group of people who are indigenous to the region which is now present-day Northern Sudan and southern Egypt. They originate from the early Sub-Saharan African inhabitants of the central Nile valley, believed to be one of the earliest cradles of civilization.
| Please note: Like most Tomb paintings, this painting has been repainted: ostensibly for "Repair" of damaged parts. But as we all know, the Albinos and their Mulattoes have a habit of "Repairing" such that the subject reappears with Albino features and light skin. |
Nubian Weddings
ABSTRACT
The area known today as Sudan may have been the scene of pivotal human evolutionary events, both as a corridor for ancient and modern migrations, as well as the venue of crucial past cultural evolution. Several questions pertaining to the pattern of succession of the different groups in early Sudan have been raised. To shed light on these aspects, ancient DNA (aDNA) and present DNA collection were made and studied using Y-chromosome markers for aDNA, and Y-chromosome and mtDNA markers for present DNA. Bone samples from different skeletal elements of burial sites from Neolithic, Meroitic, Post-Meroitic and Christian periods in Sudan were collected from Sudan National Museum. Fourteen samples were females and 19 were males. To generate Y-chromosome specific haplogroups A-M13, B-M60, F-M89 and Y Alu Polymorphism (YAP) markers, which define the deep ancestral haplotypes in the phylogenetic tree of Y-chromosome were used. Haplogroups A-M13 was found at high frequencies among Neolithic samples. Haplogroup F-M89 and YAP appeared to be more frequent among Meroitic, Post-Meroitic and Christian periods. Haplogroup B-M60 was not observed in the sample analyzed. For extant DNA, Y-chromosome and mtDNA haplogroup variations were studied in 15 Sudanese populations representing the three linguistic families in Sudan by typing the major Y haplogroups in 445 unrelated males, and 404 unrelated individuals were sequenced for the mitochondrial hypervariable region.
Y-chromosome analysis shows Sudanese populations falling into haplogroups A, B, E, F, I, J, K, and R in frequencies of 16.9, 8.1, 34.2, 3.1, 1.3, 22.5, 0.9, and 13% respectively. Haplogroups A, B, and E occur mainly in Nilo-Saharan speaking groups including Nilotics, Fur, Borgu, and Masalit; whereas haplogroups F, I, J, K, and R are more frequent among Afro-Asiatic speaking groups including Arabs, Beja, Copts, and Hausa, and Niger-Congo speakers from the Fulani ethnic group.
For mtDNA analysis, a total of 56 haplotypes were observed, all belonging to the major sub-Saharan African and Eurasian mitochondrial macrohapolgroups L0, L1, L2, L4, L5, L3A, M and N in frequencies of 12.1, 11.9, 22, 4.2, 6.2, 29.5, 2, and 12.2% respectively. Haplogroups L6 was not observed in the sample analyzed. The considerable frequencies of macrohaplogroup L0 in Sudan is interesting given the fact that this macrohaplogroup occurs near the root of the mitochondrial DNA tree. Afro-Asiatic speaking groups appear to have sustained high gene flow form Nilo-Saharan speaking groups. Accordingly, through limited on number of aDNA samples, there is enough data to suggest and to tally with the historical evidence of the dominance by Nilotic elements during the early state formation in the Nile Valley, and as the states thrived there was a dominance by other elements particularly Nuba/Nubians.
In Y-chromosome terms this mean in simplest terms introgression of the YAP insertion (haplogroups E and D), and Eurasian Haplogroups which are defined by F-M89 against a background of haplogroup A-M13. The data analysis of the extant Y-chromosomes suggests that the bulk of genetic diversity appears to be a consequence of recent migrations and demographic event mainly from Asia and Europe, evident in a higher migration rate for speakers of Afro-Asiatic as compared to the Nilo-Saharan family of languages, and a generally higher effective population size for the former. While the mtDNA data suggests that regional variation and diversity in mtDNA sequences in Sudan is likely to have been shaped by a longer history of in-situ evolution and then by human migrations form East, west-central and North Africa and to a lesser extent from Eurasia to the Nile Valley
By Verena J. Schuenemann, Alexander Peltzer, Beatrix Welte, W. Paul van Pelt, Martyna Molak, Chuan-Chao Wang, Anja Furtwängler, Christian Urban, Ella Reiter, Kay Nieselt, Barbara Teßmann, Michael Francken, Katerina Harvati, Wolfgang Haak, Stephan Schiffels & Johannes Krause.
Abstract
Egypt, located on the isthmus of Africa, is an ideal region to study historical population dynamics due to its geographic location and documented interactions with ancient civilizations in Africa, Asia and Europe. Particularly, in the first millennium BCE Egypt endured foreign domination leading to growing numbers of foreigners living within its borders possibly contributing genetically to the local population. Here we present 90 mitochondrial genomes as well as genome-wide data sets from three individuals obtained from Egyptian mummies. The samples recovered from Middle Egypt span around 1,300 years of ancient Egyptian history from the New Kingdom to the Roman Period. Our analyses reveal that ancient Egyptians shared more ancestry with Near Easterners than present-day Egyptians, who received additional sub-Saharan admixture in more recent times. This analysis establishes ancient Egyptian mummies as a genetic source to study ancient human history and offers the perspective of deciphering Egypt’s past at a genome-wide level
The study was able to measure the mitochondrial DNA of 90 individuals, and it showed that the mitochondrial DNA composition of Egyptian mummies has shown a high level of affinity with the DNA of the populations of the Near East. A shared drift and mixture analysis of the DNA of these ancient Egyptian mummies shows that the connection is strongest with ancient populations from the Levant, the Near East and Anatolia, and to a lesser extent modern populations from the Near East and the Levant. In particular the study finds "that ancient Egyptians are most closely related to Neolithic and Bronze Age samples in the Levant, as well as to Neolithic Anatolian and European populations".
Genome-wide data could only be successfully extracted from three of these individuals. Of these three, the Y-chromosome haplogroups of two individuals could be assigned to the Middle-Eastern haplogroup J, and one to haplogroup E1b1b1 common in North Africa. The absolute estimates of sub-Saharan African ancestry in these three individuals ranged from 6 to 15%, which is significantly lower than the level of sub-Saharan African ancestry in the modern Egyptians from Abusir, who "range from 14 to 21%."( When using East African admixed population as reference) The study's authors cautioned that the mummies may be unrepresentative of the Ancient Egyptian population as a whole, since they were recovered from the northern part of Egypt. Overall the mummies studied were closer genetically to Near Eastern people than the modern Egyptian population, which has a greater proportion of genes coming from sub-Saharan Africa after the Roman period.
The data suggest a high level of genetic interaction with the Near East since ancient times, probably going back to Prehistoric Egypt: "Our data seem to indicate close admixture and affinity at a much earlier date, which is unsurprising given the long and complex connections between Egypt and the Middle East. These connections date back to Prehistory and occurred at a variety of scales, including overland and maritime commerce, diplomacy, immigration, invasion and deportation". Blood typing and ancient DNA sampling on Egyptian mummies is scant. However, blood typing of Dynastic period mummies found their ABO frequencies to be most similar to that of modern Egyptians.
Separately: The remains of a Canaanite Nobelman from 1,300 B.C. was found in Tel Shadud, an archaeological mound in the Jezreel valley of the Northern District of what is Israel today: he was of Y-haplogroup R1b.
By Alice Baghdjian
LONDON – Up to 70% of British men and half of all Western European men are related to the Egyptian Pharaoh Tutankhamun, geneticists in Switzerland said. Scientists at Zurich-based DNA genealogy centre, iGENEA, reconstructed the DNA profile of the boy Pharaoh, who ascended the throne at the age of nine, his father Akhenaten and grandfather Amenhotep III, based on a film that was made for the Discovery Channel.
| This Newspaper is implying that R1b is a uniquely Albino Y-haplogroup. This is the kind of delusional lie we at RHWW always warn against. And that is why we show all of those Black R1b people, and explain about the Black people where the haplogroup evolved. Worst yet, these delusional Albinos fantasies are often encouraged by Albino scientists and famous Albino institutions - in this case the Discovery Channel. Of course the essential point is "Albino": whether it be the lowest White trash or the powerful board chairman, Albinos uniformly need the lies of past greatness to rationalize the crimes they committed to gain control of a Black World that still struggles with taking Albinos seriously. Of course that comes under the heading of "Dumb Nigger": even a child can kill. |
The results showed that King Tut belonged to a genetic profile group, known as haplogroup R1b1a2, to which more than 50% of all men in Western Europe belong, indicating that they share a common ancestor. Among modern-day Egyptians this haplogroup contingent is below 1%, according to iGENEA. “It was very interesting to discover that he belonged to a genetic group in Europe — there were many possible groups in Egypt that the DNA could have belonged to,” said Roman Scholz, director of the iGENEA Centre. The non-scientific Albino world wanted to run with the delusional lie, but Roman Scholz was trying distance himself from the lie, while not actually disagreeing with the idiot Albinos.
Which is really an Osiris Mask?Clearly Albinos make it up as they go! |
| Click for Realhistoryww Home Page |